Development of Continuous Validation Model on Standard Codes Mapping for Multi-Institutional Collaborative Data-Driven Medical Study
2 Medical Information Centre, Kyushu University Hospital, Fukuoka, Japan, Email: yamashita-takanori@meti.go.jp
3 Department of Clinical Chemistry and Laboratory Medicine, Kyushu University Hospital, Fukuoka, Japan, Email: thotta@med.kyushu-u.ac.jp
4 Head office, Tokushukai Information System Incorporated, Osaka, Japan, Email: fujimura.y-tisi@ac.jp
5 Department of Medical Informatics, Hamamatsu University Hospital, Shizuoka, Japan, Email: kimura@mi.hama-med.ac.jp
6 Medical IT Centre, Tohoku University Hospital, Sendai, Japan, Email: m.nakayama@tohoku.ac.jp
7 Department of Healthcare Information Management, The University of Tokyo Hospital, Tokyo, Japan, Email: ohe.k-tky@umin.ac.jp
8 Department of Pharmacy, NTT Medical Centre Tokyo, Tokyo, Japan, Email: orii-tky@umin.ac.jp
9 Department of Laboratory Medicine, Saga University Hospital, Saga, Japan, Email: eiz.sueoka@med.su.jp
10 Department of Medical Informatics, Chiba University Hospital, Chiba, Japan, Email: suzuki@ho.chiba-u.ac.jp
11 Department of Medical Informatics, Kagawa University Hospital, Kagawa, Japan, Email: medinfo@med.kagawa-u.ac.jp
Received: 17-Jul-2020 Accepted Date: Aug 05, 2020 ; Published: 12-Aug-2020
Citation: Park J, Yamashita T, Takada A, Hotta T, Nojiri C, Izukura R, et al. (2020). Development of Continuous Validation Model on Standard Codes Mapping for Multi-institutional Collaborative Data-Driven Medical Study. EJBI. 16(3): 10-19.
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact submissions@ejbi.org
Abstract
Background: The Medical Information Database Network (MID-NET) is a national project that promotes effective safety measures for the active surveillance of drug safety assessments through pharmacoepidemiological methods, using real-world data in Japan. The MID-NET contains the data of approximately 5.05 million patients (as of December 2019) across 10 medical institutions, including 23 hospitals. One of the most important conditions for conducting pharmacoepidemiological research using multiple medical databases is to systematically verify of data standardization.
Objectives: To evaluate the effect of improving the accuracy of standard data quality control by the development of a validation model for standard code mapping in multiple medical information databases.
Methods: We established the standard code mapping validation center at one of the cooperating medical institutions of the MID-NET that could collect and manage information about the standard code interoperability. Additionally, we used the mapping table for the four standard codes, including the Japan Laboratory Test Standard Code, 10th Revision (JLAC10) code were collected from MID-NET cooperating institutions, and the accuracy of the mapping table was evaluated.
Results: The observed four standard codes mapping ratio between institutions varied from >2,000 to <100. Moreover, the accuracies of standard codes were not standardized. We used a centralized standard code mapping validation model to provide feedback for standardizing JLAC-10 for each institution and meaningful differences between institutions were improved. Conclusions: The developed model visualized information differences and improved the data quality between multiple medical institutions.
http://www.oajournal.org/
http://www.journalsres.org/
http://www.journalsres.com/
http://www.journalsoa.org/
http://www.journalsoa.com/
http://www.journalsci.org/
http://www.journalres.org/
http://www.journalres.com/
http://www.journaloa.org/
http://www.journalinsights.org/
http://www.jpeerreview.org/
http://www.imedresearch.com/
http://www.imedpubjournals.com/
http://www.imedpubjournal.org/
http://www.imedjournals.org/
http://www.peerreviewedjournal.org/
http://www.peerjournals.org/
http://www.peerjournals.com/
http://www.sciencesinsight.org/
http://www.scholarresearch.com/
http://www.scholarres.org/
http://www.nutritionres.com/
http://www.gastroinsights.org/
http://www.pathologyinsights.org/
http://www.echemistry.org/
http://www.echemcentral.com/
http://www.chemistryres.com/
http://www.biochemresearch.org/
http://www.biochemjournals.com/
http://www.ebusinessjournals.org/
http://www.businessjournals.org/
http://www.peerjournal.org/
http://www.oajournalres.com/
http://www.alliedres.org/
http://www.alliedjournals.org/
http://www.alliedjournal.org/
http://www.scientificres.org/
http://www.scientificres.com/
,
https://www.mongoliannutrition.com/
https://www.nsbmb.com/
https://www.arabspp.org/
https://www.arabianmultidisciplinary.com/
https://www.italystemcell.com/
https://www.traditional-medicine.org/
https://www.episportsmedicine.org/
https://www.worldmedicalassociation.org/
https://www.silaeitaly.com/
https://www.ceramicsmedicine.org/
https://www.isaddictionmedicine.org/
https://www.europeanbionetwork.com/
https://www.aarsecp.com/
https://www.edycseg.org/
https://www.europeanneurology.org/
https://www.clinicaldermepi.com/
https://www.cardiac-society.com/
https://www.psychologicalassociation.org/
https://www.indian-psychology.com/
https://www.mongoliancardiology.org/
https://www.pediatricssociety.com/
https://www.cocrt.org/
https://www.european-aesthetic.com/
https://www.sohnsb.org/
Keywords
Medical data management; Medical information database; Data interoperability; Data quality assurance; Real-world data
Introduction
Data standardization and clearly defined data validation methods are important topics in the data-driven technology field. Recently, with the widespread implementation of “Real-World Data” (RWD) in data-driven medical studies (DDMS), there is increasing interest in the use of these high-quality electronic medical records and information-sharing interoperability resources to enhance the study of serious Adverse Drug Reactions (ADRs) [1-3]. Furthermore, harmonizing multiple databases in DDMS may increase pharmacoepidemiological research efficiency in identifying ADR cases that use potential evidencebased medicine to quantitatively evaluate both short-term and long-term benefits [2,4-6].
A typical example is the U.S. Food and Drug Administration (FDA) “Sentinel Initiative,” which established the “Mini- Sentinel,” a nationally distributed electronic system to monitor the pharmaceutical safety of FDA-regulated medical products [7-9]. In order to begin building large-scale, multiple health care databases at the national level in Japan, the National Database of Health Insurance Claims and Specific Health Checkups of Japan (NDB) were constructed in the fiscal year 2009 [10]. The database includes the data of almost all patients who received medical care services under the national health insurance system in Japan, which covers more than 126 million people and 1.9 billion claims annually [11]. Furthermore, along with health and medical information computerization and progress, large databases using the Standardized Structured Medical Information eXchange2 (SS-MIX2) system has been established [12,13].
For example, the Medical Information Database Network (MIDNET) project (previously known as the “Japanese Sentinel Project”), based on the SS-MIX2 system, has established new RWD from multiple medical institutions in Japan [14,15]. The Ministry of Health, Labor and Welfare of Japan established this project as a scientific approach to determining safety measures for ADRs to pharmaceuticals. The Pharmaceuticals and Medical Devices Agency (PMDA) started full-scale operation of this project in the fiscal year 2018. The database in this project consists of RWD collected from approximately 5.05 million patients at 10 medical institutions, including 23 hospitals in Japan (Chiba University Hospital, Hamamatsu University Hospital, Kagawa University Hospital, four hospitals from the Kitasato Institute Group, Kyushu University Hospital, Tohoku University Hospital, 10 hospitals from the Tokushukai Medical Group, and two hospitals from the NTT Hospital Group, Saga University Hospital, and the University of Tokyo Hospital). This project aims to promote effective safety measures to minimize the risks and maximize the benefits of drugs, through pharmacoepidemiological methods, using RWD.
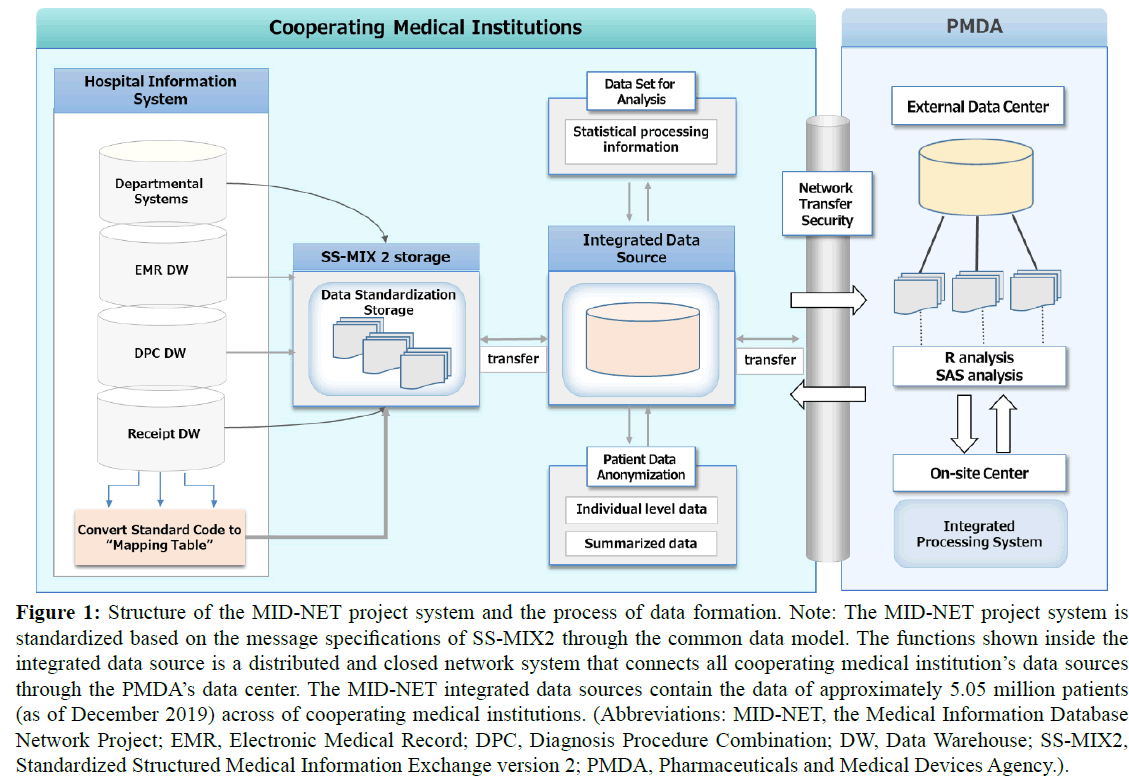
In this project, the database of medical information from the Hospital Information Systems (HISs) of cooperating medical institutions is stored in the MID-NET integrated Data Source (DS) via the SS-MIX2. As shown in Figure 1, the MID-NET integrated DS, installed at each cooperating medical institution, is linked to a PMDA’s on-site data center based on a distributed infrastructure network system. This distributed integrated DS system is used for the stored database network. Regular updating of stored data from clinical practice is done so that the data is up-to-date. Additionally, it is composed of standardized database systems, retrieved from the electronic health records of cooperating institutions, to analyze and evaluate ADRs. Furthermore, patient individual-level data are automatically anonymized to protect privacy information (name, address, and residential postal code) and sent to PMDA for integrated analysis.
Figure 1: Structure of the MID-NET project system and the process of data formation. Note: The MID-NET project system is standardized based on the message specifications of SS-MIX2 through the common data model. The functions shown inside the integrated data source is a distributed and closed network system that connects all cooperating medical institution’s data sources through the PMDA’s data center. The MID-NET integrated data sources contain the data of approximately 5.05 million patients (as of December 2019) across of cooperating medical institutions. (Abbreviations: MID-NET, the Medical Information Database Network Project; EMR, Electronic Medical Record; DPC, Diagnosis Procedure Combination; DW, Data Warehouse; SS-MIX2, Standardized Structured Medical Information Exchange version 2; PMDA, Pharmaceuticals and Medical Devices Agency.).
The distributed MID-NET integrated DS system includes 11 types of currently available standard codes when converting data from the HIS information to a mapping table based on local codes, such as including the medical examination history code (including admission and discharge data); diagnostic orders code; discharge summary code; International Statistical Classification of Diseases and Related Health Problems, 10th Revision (ICD- 10) code by the World Health Organization (WHO) [16]; Japan pharmaceutical permanent reference codes (called the HOT code) of prescription and injection orders/execution data [17], Japan national health insurance drug codes (called the YJ code) of prescription and injection orders/execution data [18,19]; radiographic inspection data; physiological laboratory data; therapeutic drug monitoring data; bacteriological test data; and Japan Laboratory Test Standard Code, 10th Revision (called the JLAC-10) code. The JLAC-10 is based on the Health Level 7 International Standards and was established using the SS-MIX2 system [20]. The use of such data is expected to help establish standard databases such as those for local medical information linkage and DDMS in the future.
Specifically, distributed integrated DS systems, such as MIDNET, integrate DS-aggregated clinical data from collaborating medical institutions for pharmacoepidemiological analysis; thus, conversion (mapping) from local hospital codes to a standard code is essential in the database [21,22]. Because quality assurance of the mapping tables affects SS-MIX 2 storage of standardized data, it is important not only for the MID-NET project and but also for the construction of medical information databases for the secondary use of large-scale medical information. However, the MID-NET validation project prioritized the management of the medical institutions’ local codes associated with medical services, which resulted in delays in mapping coding or omissions in standardized data [23]. Because these factors are temporary and occur continuously and unexpectedly at each institution, integrated management is extremely difficult and results in poor data quality.
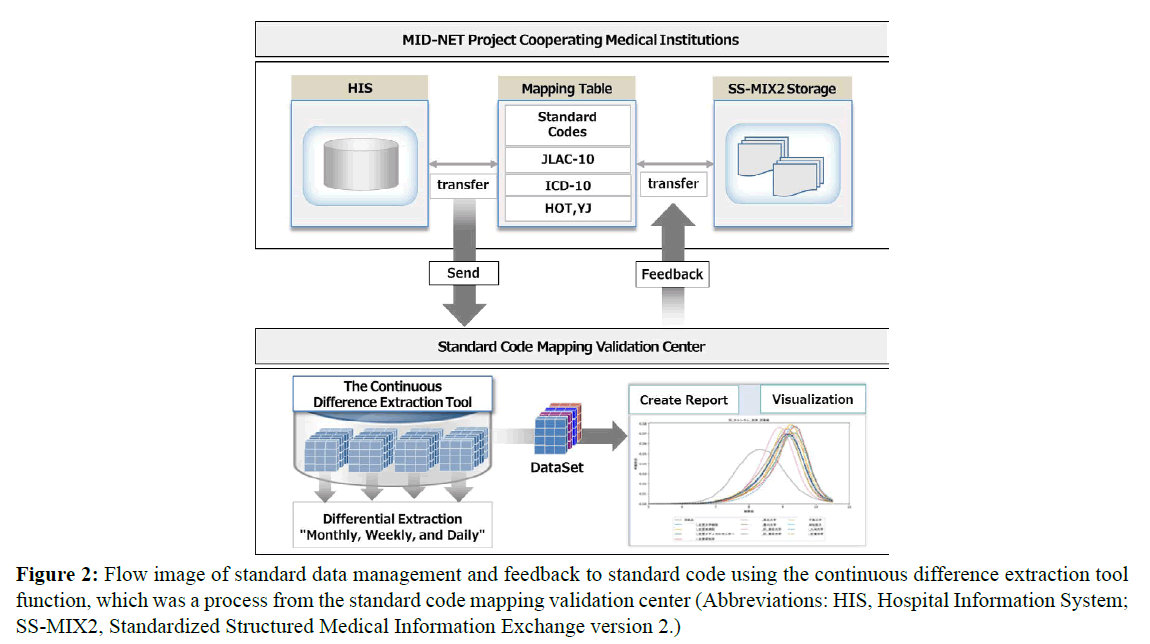
Therefore, the MID-NET project expert meeting suggested the need for a specialized organization to manage multiple database links and data quality uniformly, at a centralized organization level, including providing mapping table maintenance and management. Based on the preceding discussion, the research group engaged in this study received support from the Japan Agency for Medical Research and Development (AMED) to establish the standard code mapping validation center at the Kyushu University Hospital that can maintain and manage the standard codes interoperability of the MID-NET projectcooperating medical institutions. Moreover, the group developed a continuous difference extraction tool (Version 1) for central management based on a system consisting of 11 types of standard codes; the group introduced this tool in three MID-NET projectcooperating medical institutions (Figure 2).
Figure 2: Flow image of standard data management and feedback to standard code using the continuous difference extraction tool function, which was a process from the standard code mapping validation center (Abbreviations: HIS, Hospital Information System; SS-MIX2, Standardized Structured Medical Information Exchange version 2.)
Objectives
This study aimed to establish and evaluate the effect of improving data quality control on secondary use of clinical data by providing a centralized standard code mapping validation model that improves quality accuracy of standard code interoperability and consistency from hospital information systems of multiple medical institutions, using the MID-NET project network. There was a specific focus on understanding and developing the continuous difference extraction tool’s effectiveness for data standardization management with high-performance quality. The main contributions of this study included the following: (1) empirical evidence for a centralized standard data mapping validation model to support the need for data standardization in data-driven clinical epidemiological studies using large-scale databases and (2) methodological suggestions for multiple-site data standardization.
Methods
Study design and data source
This study focused on four of major standard codes, namely, the JLAC-10, HOT, YJ, and ICD-10 codes, and evaluated (1) the mapping ratios of standard codes, (2) the ratio of JLAC-10 codes matching among medical institutions, and (3) the continuous status of standard codes in terms of quality management among institutions. Therefore, mapping tables were requested from nationwide MID-NET-cooperating medical institutions (23 hospitals of 10 medical institutions). Among these, the mapping tables were collected from nine medical institutions (including 18 hospitals) through the standard code mapping validation center.
Evaluation of the matching ratio with the JLAC-10 (224 items) adopted by the MID-NET Project
First, the 224 items of the JLAC-10 adopted in the MID-NET project were used as a “key codes” to evaluate the accuracy of the matching ratio of clinical laboratory standard codes among the cooperating medical institutions. The PMDA adopted the JLAC-10 classification 224 items (as of September 2017) because it had the highest priority for clinical examinations in terms of clinical laboratory test frequency and safety measures as items to be considered for data quality defects before full-scale operation of MID-NET project. Thus, the matching ratio between mapping tables was assessed using the 224 items of the JLAC-10 as a positive specifying key code; 90% of all routine clinical laboratory tests are included in the MID-NET project key code (e.g., hemoglobin, urea nitrogen, peripheral blood, and creatinine).
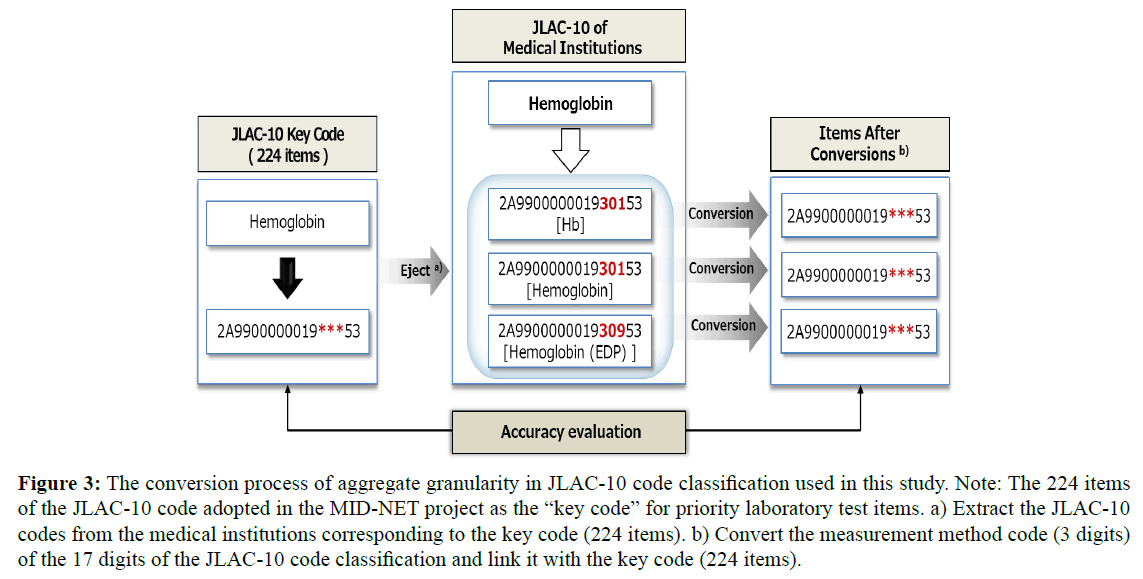
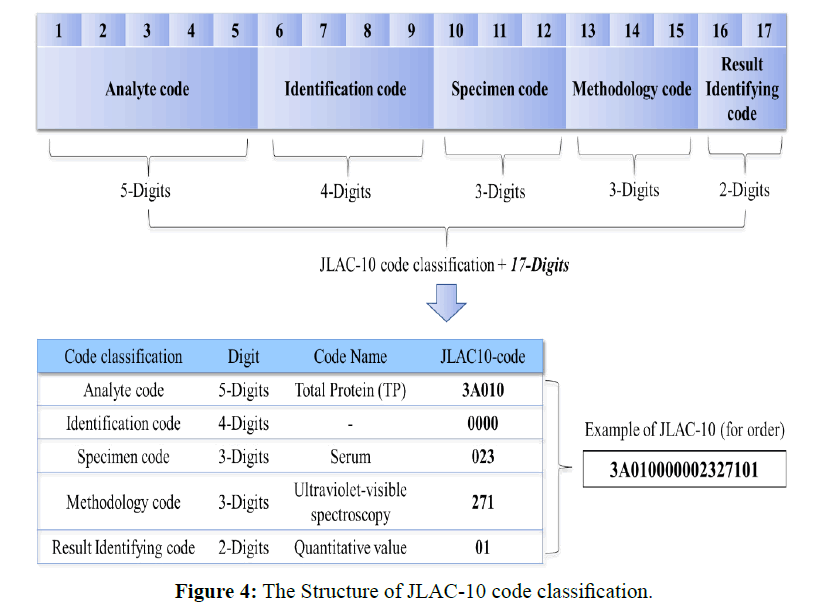
Next, Figure 3 shows the conversion process of aggregate granularity in JLAC-10 code classification used in this study. The granularity adjustment table was created using for clinical laboratory test mapping table among the collaborating medical institutions in terms of obtaining accurate coding results respectively in JLAC-10 code classification. The JLAC-10 contains five segments (17 digits): analyte code (5 digits), identification code (4 digits), material code (3 digits), measurement method code (3 digits), and result identification code (2 digits), and its structure is illustrated in Figure 4. It is based on international laboratory tests coding standards such as Logical Observation Identifier Names and Codes (LOINC). For these segments (17 digits), the code granularity was adjusted, such that the codes were linked 1:1 without regard for the measurement method code (3 digits). The measurement method code (3 digits) was omitted because the JLAC-10 contains factors that cause medical institute-specific variations in the interpretation of each component, with a strong tendency for fluctuations in the measurement method segment.
Figure 3: The conversion process of aggregate granularity in JLAC-10 code classification used in this study. Note: The 224 items of the JLAC-10 code adopted in the MID-NET project as the “key code” for priority laboratory test items. a) Extract the JLAC-10 codes from the medical institutions corresponding to the key code (224 items). b) Convert the measurement method code (3 digits) of the 17 digits of the JLAC-10 code classification and link it with the key code (224 items).
Finally, the presence of JLAC-10 corresponding to the 224 items was examined using the mapping table. Additionally, when the clinical laboratory test codes were updated, and multiple JLAC- 10 items were linked to the same local code, the JLAC-10 was counted as one case.
Evaluation of the introduced continuous difference extraction tool for data standardization management
This study evaluated the real-time status of data standardization management, after initially introducing the continuous validation model at three MID-NET cooperating medical institutions, to identify differences in the JLAC-10 codes between institutions from November 2018 to March 2019. This difference extraction tool for data standardization management in realtime automatically transfers “differential” information that is extracted on a monthly, weekly, and daily basis for 11 types of standard codes, including the JLAC-10, HOT, YJ, and ICD-10 codes, delivered to the standard code mapping validation center.
Here, “difference” indicates an “event” in which a standard code was newly added or changed according to a unique local laboratory test code in a medical institution’s mapping table. If there were no events to local codes in HIS, the difference information is displayed in the “Null” state. These results provided feedback on the real-time status of standard data quality management to forms among the medical institutions. During this process, the study also used the JLAC-10 key codes (224 items) to evaluate the accuracy of the differences extracted by the tool: (1) an exact match ratio with the key codes (224 items), and (2) the improvement ratio of before and after feedback on the accuracy of the JLAC-10 coding by the mapping validation center.
Ethical Considerations
This study has been reviewed and approved by the Kyushu University medical ethics committee (reference number: 2019-021).
Results
The mapping ratio of the four standard codes in the MIDNET of cooperative medical institutions
Table 1 presents the mapping ratio of the four standard codes in the mapping table: At least 2,000 items of the JLAC-10 were mapped in the order of cooperating institutions (A), (G), and (F) and the entire hospital group (I). However, fewer than 100 items on JLAC-10 were mapped for the institutions (E), (C), and (D). In particular, in the institution (D), only 34 items on JLAC-10, of the total 4,514 unique local laboratory codes associated with all clinical laboratory tests, were mapped on the mapping table.
| Aggregate codes | MID-NET Cooperating Medical Institutions | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| (A) | (B) | (C) | (D) | (E) | (F) | (G) | (H) | (I)† | |
| Clinical laboratory test | |||||||||
| Laboratory test local code | 5,704 | 5,671 | 4,763 | 4,514 | 3,610 | 2,788 | 2,777 | 2,256 | 19,066 |
| JLAC-10 code | 3,186 | 682 | 43 | 34 | 87 | 2,424 | 2,777 | 1,563 | 17,306 |
| Pharmaceutical (prescription) | |||||||||
| Pharmaceutical local code | 4,365 | 5,883 | 6,263 | Null | 3,528 | 43,387 | 5,769 | 4,135 | Null |
| HOT code | 2,347 | Null | 40 | Null | 2,826 | 16,731 | 3,077 | 4,131 | Null |
| YJ code | 4,365 | 2,181 | 6,263 | Null | 3,528 | 41,158 | Null | Null | Null |
| Pharmaceutical (injection) | |||||||||
| Pharmaceutical local code | 1,947 | 3,132 | 2,359 | Null | 1,627 | 1,408 | 2,336 | 4,234 | Null |
| HOT code | 1,073 | Null | 10 | Null | 2 | 1,408 | 900 | 4,230 | Null |
| YJ code | 1,947 | 1,102 | 2,359 | Null | 1,627 | 1,312 | Null | Null | Null |
| Disease name | |||||||||
| Disease name local code | 30,681 | 31,693 | 33,799 | Null | 30,393 | 28,540 | Null | Null | Null |
| ICD-10 code | 30,653 | 28,289 | 33,632 | Null | 28,656 | 26,017 | Null | Null | Null |
Table 1: The number of local codes and standard codes in the mapping table. Abbreviations: JLAC-10 code, Japan Laboratory Test Standard Code, 10th revision; HOT code, Japan pharmaceutical permanent reference code; YJ code, Japan national health insurance drug code; ICD-10 code, International Classification of Diseases Code, 10th revision code by the World Health Organization (WHO). †Total number of mapped codes of the entire hospital organization in one hospital group.
Regarding pharmaceuticals, four institutions, (A), (C), (E), and (F), out of nine medical institutions had both HOT and YJ codes mapped, including prescriptions and injection items for pharmaceuticals. In particular, the ratio of YJ code was higher than for the HOT code. The ICD-10 codes showed a high mapping ratio of more than half of all disease name local codes.
The matching ratio for the JLAC-10 (224 items) between medical institutions
Table 2 shows the matching ratio using the JLAC-10 (224 items) as a “key codes” in the mapping tables that linked nine medical institutions. The matching ratio linked to the key codes (224 items) were 84.8% for the institution (A), 79.5% for the institution (G), 62.1% for the institution (F), and 59.4% for the institution (B). Also, more than two-thirds (50%) of the total codes matched between the key code and the four medical institutions. By contrast, the five medical institutions had a matching ratio of less than 50% for the key codes, including at the institutions (I) (48.2%) and (H) (39.7%). Furthermore, among the mapped codes for the institution (C), none matched the key codes of the JLAC-10. Of the key code (224 items), an average of 99.7 (44.5%) was matched among all nine medical institutions, whereas 124.3 (55.5%) did not match in any of the institutions.
| Medical | Comparison of JLAC-10 code vs JLAC-10 key code (224 items) | ||||
|---|---|---|---|---|---|
| Institutions | Match (n = 224) | Mismatch (n = 224) | |||
| N | % | N | % | ||
| (A) | 190 | 84.8 | 34 | 15.2 | |
| (B) | 133 | 59.4 | 91 | 40.6 | |
| (C) | 0 | 0 | 43 | 19.2 | |
| (D) | 25 | 11.2 | 199 | 88.8 | |
| (E) | 35 | 15.6 | 189 | 84.4 | |
| (F) | 139 | 62.1 | 85 | 37.9 | |
| (G) | 178 | 79.5 | 46 | 20.5 | |
| (H) | 89 | 39.7 | 135 | 60.3 | |
| (I)§ | 108 | 48.2 | 116 | 51.8 | |
Table 2: The matching ratio of JLAC-10 code between each cooperating medical institutions and the JLAC-10 key code (224 items). Note: The local laboratory codes associated with the JLAC-10 Key code (224 items) were present in the mapping table, but when the JLAC-10 code has not been coded or the JLAC-10 code has been coded but does not match 224 items, it was counted to be a mismatch. §One of 10 hospitals in one hospital group.
Specifically, the code that showed the most mismatch with the key codes was an allergen-specific test-related code, corresponding to the analysis target code of the 17-digit classification of the JLAC- 10. Furthermore, many items had fluctuations in laboratory test standards, such as the measurement method and measurement amount (e.g., urine test, blood erythroblast test, urine specific gravity test, uric acid test, and bodyweight test). Additionally, there were coding errors in the element code of laboratory tests; in such cases, the JLAC-10 of 17-digit classification contained the specific characteristics such as “#,” “*,” or “☆.” For example, as shown in Table 1, for the medical institution (I), 5.5% of the 17,306 items on JLAC-10 in the mapping table were coded with these unique specific character symbols. In such cases, it was confirmed by a visit survey in the mapping validation Centre that the character words unique to the JLAC-10 were coded when the clinical laboratory test outsourcing contractor was updated.
Experimental results of difference detection using the continuous difference extraction tool and validation of mapping accuracy and consistency with key codes (224 items) based on the feedback
The continuous difference extraction tool was introduced at the medical institution (A), which had >2,000 of the JLAC-10 items on the mapping table, and at medical institutions (C) and (E), which each had <100 of the JLAC-10. Table 3 shows the monthly results of outputting the differences in events detected continuously by the difference extraction tool. There were 4,842 differences in items detected between December 2018 and January 2019 from the laboratory test items of the institution (C). Among these, there were no items to which the JLAC-10 was mapped, and all differences in events were detected continuously, using unique local laboratory test codes without the JLAC-10. During the same period, 3,491 items in differences were detected for the institution (E). Among these, there were 45 items in differences to which the JLAC-10 were mapped, and 3,446 items in differences detected without the JLAC-10.
| Medical | Time series of monthly difference extraction results | Matching ratio | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Institutions | 2018/12 | 2019/01 | Feed | 2019/02 | with Key code | ||||
| JLAC-10 | JLAC-10 | back | JLAC-10 | (224 items) | |||||
| Presence | None | Presence | None | Presence | None | Nc) | % | ||
| (only local code) | (only local code) | (only local code) | |||||||
| (A) | 0 | 12 | 0 | 7 | ➡ | 9 | 0 | 190 | 84.8 |
| (n = 12) | (n = 7) | (n = 9) | (N/Ad,e) | ||||||
| (C) | 0 | 4837 | 0 | 5 | ➡ | 224 | 0 | 224 | 100.0 (+100.0d) |
| (n = 4837) | (n = 5) | (n = 224) | |||||||
| (E) | 45 | 3446 | 0 | 0 | ➡ | 169 | 11 | 200 | 89.3 |
| (n = 3491) | (n = 0) | (n = 180) | (+82.5d) | ||||||
Table 3: The matching ratio with the key code (224 items) provided as feedback using the continuous difference extraction tool. Note: The standard code mapping validation center summarized the mapping status of JLAC-10 code until January 2019, and showed the improvement rate for the accuracy of the JLAC-10 code mapping status and code granularity after feedback to three medical institutions. c) The number of items matched with the key code (224 items) in the new mapping table after feedback. d) The rate of improvement in the ratio of JLAC-10 code that matches with the key code before and after feedback. e) Not applicable.
A list of these differences where the JLAC-10 codes were not mapped was generated on the mapping validation center, and feedback was provided to the three institutions (A), (C), and (E) in a report format. After that, the newly detected difference data was scrutinized again using the continuous difference extraction tool in February 2019. It detected a difference on February 10, 2019, for the institution (C). It was confirmed that there were 224 items with JLAC-10 in the local laboratory test code newly coded. For the institution (E), 180 differences were detected that were newly assigned on February 17, 2019. Of these 180 differences, it was confirmed that 169 items were newly coded with JLAC-10 codes.
Next, the accuracy of the code changes made in the JLAC-10 coding of each institution after receiving feedback from the difference extraction tool was evaluated using the key codes. Of the 224 total differences detected for the institution (C), all 224 items newly assigned to JLAC-10 codes (100%) matched the key codes (224 items). For institution (E), of the 214 newly assigned items, 200 items (89.3%) matched the key codes (224 items). When our mapping validation Centre provided feedback for the institution (E), the matching ratio for the JLAC-10 to the key codes (224 items), before and after receiving feedback, improved by +82.5% compared to before the feedback. The institution (A) had a matching ratio of 84.8%, and there was no change in the granularity of the initial verification results and the JLAC-10 code.
Discussion
The data quality in medical information management in clinical practice depends on each medical institution’s systems. Consequently, standard data must be continuously reviewed via precision data management. This is an important issue because the accuracy of code mapping directly affects data quality and the ability of the system to generate valid inferences through secondary use of clinical data. However, there have been no reports of cases wherein the data quality defects that occur continuously because of multiple databases are centrally managed on a computer system. Therefore, this study compared the mapping ratio of the standard codes used at MID-NETcooperating medical institutions and evaluated the effect of improving data quality control by using a centralized standard code mapping validation model.
We also evaluated whether the standard code difference extraction tool for data standardization management can detect the data quality defects that occur in real-time. In particular, the continuous difference extraction tool, using the centralized validation model were examined, providing each medical institution with feedback on differences regarding the mapping validation center detected continuously; through this process, the effects that lead to improved data quality between multiinstitutional databases could be identified. Also, the study provided an overall view of MID-NET data quality management, including past time series data, to simultaneously improve the data interoperability between databases.
As shown in Tables 1 and 2, this study revealed that the standard codes matching ratio varied within medical institutions and that the underlying factors for these variations might be due to included differences in specifications among the HIS department systems, differences between electronic medical record vendors, and differences in the management structures at individual institutions. More specifically, a mismatch was found in the consistency of key codes (224 items) with JLAC-10 between medical institutions. The reasons for the inconsistencies in JLAC- 10 in the mapping table at each institution can be categorized as follows: (1) the low JLAC-10 coding ratio in the mapping table; (2) differences between JLAC-10 and key codes (224 items) in operation at institutions; (3) the history of revisions of local codes related to unmanaged laboratory tests (changes in testing equipment, changes in outsourcing companies, etc.); and (4) unevaluated coding errors and mistakes when registering a medical records management department’s JLAC-10.
In a mapping table, there were also medical institutions with “Null” status for the HOT/YJ codes and ICD-10 codes among the standard codes. This may be attributed to the update of the mapping table, which is conducted irregularly and simultaneously additions and changes at the clinical site. Moreover, in the case of electronic medical records, which are established for each institution, all patient information is included, but it is difficult to continuously integrate and convert to standard code because it is entered in natural language rather than structured and formalized language. Thus, it is presumed that the mapping ratio and consistency may be affected following the update based on local codes. The underlying reason appears to be that mapping table management is performed by the medical institutions concerned, and in practice, a time lag occurs for the standard code to update. These factors have a direct or indirect effect on all other latent constructs in data standardization. However, although these factors represent correlations that are consistent with the hypothesized causations, relationships comparing quality differences between the mapping tables should consider actual in-hospital system proof of causation. Additionally, because only the unique local laboratory test codes without the standard code are used in clinical practice, a mechanism that simultaneously, continuously, and correctly assigns the standard code is necessary.
This study found that understanding the frequency of standard code updates at each institution in real-time continuously could derive sources of discrepancies related to the mapping tables’ accuracy. Furthermore, this study confirmed that by using the continuous difference extraction tool, situations where irregularly differences were noted in JLAC-10 codes could be visualized; these corresponded to differences detected as the local laboratory codes’ new additions and to changes at the clinical site. As a result, it was possible to evaluate the status of erroneous standard coding and the status associated with the local laboratory test codes unique to each medical institution in coding the JLAC-10. The study also confirmed more meaningful differences in the mapping table of the distribution of JLAC-10, because the accuracy of code mapping directly affects data quality. As shown in Table 3, when our mapping validation center provided feedback to the institutions (C) and (E), the ratio of the match between MID-NET project key code (224 items) and JLAC-10 after feedback improved greatly the code mapping quality by comparison to before the feedback. In that case, after the feedback process, the institution (C) confirmed that there were 224 newly coded items with JLAC-10 in the local laboratory test code, and all items were consistent with the MID-NET project key code (224 items). Additionally, visualizing the information differences, providing those findings in continuously, and performing data standardization at the centralized validation center using the tool led to control of the discrepancy in the JLAC- 10 coding between medical institutions.
This study has demonstrated as its main contribution that, given the potential impact standardized data might have on the secondary use of large-scale medical information, the effort required to standardize differences in internal consistency on the standard codes can be reduced significantly, leading to improved data standardization. Furthermore, these findings show that it is possible and feasible to improve data standardization quality management by integrating and managing standard codes interoperability, using a central validation model, when valuable information is extracted from multiple-site databases, such as the MID-NET project. Although data inconsistencies were observed in the initial stage between cooperating institutions, the data quality improved dramatically following collaborative efforts between the MID-NET medical institutions, and PMDA for maintaining the quality of data interoperability management is guaranteed. The MID-NET project was successfully launched on April 1, 2018.
Limitations and Future Research
There are some limitations to this study. First, in the consistency evaluation of the JLAC-10, the measurement method code (3 digits) was omitted from the JLAC-10 code classification. However, the JLAC-10 contains factors that cause medical institute-specific variations in the interpretation of each component, with a strong tendency for fluctuations in the measurement method code, such as changes in the outsourcing laboratory test department. Second, the continuous difference extraction tool is a system to verify mapping information transferred from the HIS to the MIDNET integrated data source via SS-MIX2 standardized storage. Therefore, this study did not evaluate data that were generated continuously while the information was being transferred from the laboratory information systems to the HIS. This must be assessed by long-term, large-scale future studies. Accordingly, the study team recently introduced the continuous difference extraction tool (Version 2) to five MID-NET-cooperating institutions and is awaiting the results. Third, this study was conducted at MID-NET-cooperating medical institutions. In the future, to evaluate the modified centralized validation systems, database linkage at the multiple medical institutions that are not MID-NET project institutions will be necessary.
Conclusion
This study showed that using a centralized mapping validation model for standard data to ensure consistency with the standard code was effective in improving the data quality management system by detecting and unifying the mapping situation for the standard code in the secondary use of large-scale medical information. Additionally, we were able to use a centralized mapping validation model-based approach for the accuracy of standard data between multiple medical information databases that could improve the data interoperability. The continuous difference extraction tool, using the centralized validation model, positively affected the accuracy of standard code mapping, backed up and visualized information the data quality defects, and improved the data quality between multiple databases of medical institutions. This model is expected to be highly effective in other similar database networks, as well as MID-NET, for managing the accuracy and consistency of data standardization in an improved manner compared with other existing manually managed methods.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Acknowledgements
This research was supported by the Japan Agency for Medical Research and Development (AMED) under Grant Number 18mk0101064h0003 and 18mk0101075h0003. We appreciate the Ministry of Health, Labour and Welfare, PMDA, and the MIDNET- cooperative medical institutions for their cooperation in the research.
REFERENCES
- Kesselheim AS, Avorn J. New ”21st century cures” legislation: Speed and ease vs science. JAMA. 2017; 317(6): 581-582.
- Miguel A, Azevedo LF, Lopes F, Freitas A, Pereira AC. Methodologies for the detection of adverse drug reactions: comparison of hospital databases, chart review and spontaneous reporting. Pharmacoepidemiol Drug Saf. 2013; 22(1): 98-102.
- Wise L, Parkinson J, Raine J, Breckenridge A. New approaches to drug safety: A pharmacovigilance tool kit. Nat Rev Drug Discov. 2009; 8(10): 779-782.
- Yoon D, Park MY, Choi NK, Park BJ, Kim JH, Park RW. Detection of adverse drug reaction signals using an electronic health records database: comparison of the Laboratory Extreme Abnormality Ratio (CLEAR) algorithm. Clin Pharmacol Ther. 2012; 91(3): 467-474.
- Ramirez E, Carcas AJ, Borobia AM, Lei SH, Piñana E, Fudio S, et al. A pharmacovigilance program from laboratory signals for the detection and reporting of serious adverse drug reactions in hospitalized patients. Clin Pharmacol Ther. 2010; 87(1): 74-86.
- Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012; 13(6): 395-405.
- Behrman RE, Benner JS, Brown JS, McClellan M, Woodcock J, Platt R. Developing the sentinel system – A national resource for evidence development. N Engl J Med. 2011; 364(6): 498-499.
- Chan SL, Tham MY, Tan SH, Loke C, Foo B, Fan Y, et al. Development and validation of algorithms for the detection of statin myopathy signals from electronic medical records. Clin Pharmacol Ther. 2017; 101(5): 667-674.
- Platt R, Carnahan RM, Brown JS, Chrischilles E, Curtis LH, Hennessy S, et al. The U.S. Food and Drug Administration’s Mini-Sentinel Program: status and direction. Pharmacoepidemiol Drug Saf. 2012; 21(1): 1-8.
- Okumura Y, Sakata N, Takahashi K, Nishi D, Tachimori H. Epidemiology of overdose episodes from the period prior to hospitalization for drug poisoning until discharge in Japan: An exploratory descriptive study using a nationwide claims database. J Epidemiol. 2017; 27(8): 373-380.
- Ishimaru M, Matsui H, Ono S, Hagiwara Y, Morita K, Yasunaga H. Preoperative oral care and effect on postoperative complications after major cancer surgery. Br J Surg. 2018; 105(12): 1688-1696.
- Kimura M, Nakayasu K, Ohshima Y, Fujita N, Nakashima N, Jozaki H, et al. SS-MIX: a ministry project to promote standardized healthcare information exchange. Methods Inf Med. 2011; 50(2): 131-139.
- Matoba T, Kohro T, Fujita H, Nakayama M, Kiyosue A, Miyamoto Y, et al. Architecture of the Japan ischemic heart disease multimodal prospective data acquisition for precision treatment (J-IMPACT) system. Int Heart J. 2019; 60(2): 264-270.
- Yamada K, Itoh M, Fujimura Y, Kimura M, Murata K, Nakashima N, et al. The utilization and challenges of Japan’s MID-NET® medical information database network in postmarketing drug safety assessments: a summary of pilot pharmacoepidemiological Studies. Pharmacoepidemiol Drug Saf. 2019; 28(5): 601-608.
- Yamaguchi M, Inomata S, Harada S, Matsuzaki Y, Kawaguchi M, Ujibe M, et al. Establishment of the MID-NET® medical information database network as a reliable and valuable database for drug safety assessments in Japan. Pharmacoepidemiol Drug Saf. 2019; 28(10): 1395-1404.
- The World Health Organization (WHO). ICD-10 version (classification of diseases); 2017.
- Kaihara S, Takekuma R, Tsuchiya F. Standard master for pharmaceutical products (HOT reference number). JAMI. 2002; 22(4): 315-319.
- Akazawa M, Nomura K, Kusama M, Igarashi A. Drug utilization reviews by community pharmacists in Japan: identification of potential safety concerns through the brown bag program. Value Health Reg Issues. 2012; 1(1): 98-104.
- Ministry of Health, Labour and Welfare. List of drug prices and information on generic drugs [Japanese] from https://www.mhlw.go.jp/topics/2019/08/tp20190819-01.html.
- The Japan Society of Laboratory Medicine. The Japan laboratory accreditation cooperation (JLAC) 10. 2010.
- Khan AN, Griffith SP, Moore C, Russell D, Rosario AC, Bertolli J. Standardizing laboratory data by mapping to LOINC. J Am Med Inform Assoc. 2006; 13(3): 353-355.
- Barda AJ, Ruiz VM, Gigliotti T, Tsui FR. An argument for reporting data standardization procedures in multi-site predictive modeling: case study on the impact of LOINC standardization on model performance. JAMIA Open. 2019; 2(1): 197-204.
- Nakashima N. Pharmaceutical regulatory harmonization and evaluation research project “Practical analysis method and education on drug epidemiology research for benefits and risk assessment of pharmaceuticals using MID-NET.” The Japan agency for medical research and development (AMED) outsourced research and development results report. 2016.