Protein-protein interaction analysis of influenza a virus with the host (homosapiens) to determine the proteins involved in influenza a virus infection mechanism
K. Raj vardhan, Andhra Loyola College, India, Email: rajvardhank35@gmail.com
Received: 07-Apr-2021 Accepted Date: May 05, 2021 ; Published: 12-May-2021
Citation: Surya Teja G, Raj Vardhan K. (2021) - Protein-protein interaction analysis of influenza a virus with the host (homosapiens) to determine the proteins involved in influenza a virus infection mechanism. EJBI. 17(5)
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact submissions@ejbi.org
Abstract
Influenza A virus (IAV) infection is a serious public health problem all over the world. This virus belongs to the family Orthomyxoviridae and this is the only species of the virus occurring in the genus Alphainfluenzavirus. IAV consists of ss negative sense RNA as its genetic material and its genome comprises eight segments of viral RNA and each segment is complexed with trimeric viral polymerase proteins and nucleoprotein. IAV causes zoonotic infections in birds and severe respiratory infections in humans. The current study determines the various proteins in the biological processes of Influenza A virus in the host (Homosapiens). In this experiment, we retrieved a protein interaction network of Influenza A virus with Homosapiens. To this network cluster analysis was performed which resulted in 6 clusters. Further, gene enrichment analysis was performed for the clustered proteins using the Panther GO database and we, therefore, identified proteins that have a highly effective role in cellular processes and viral infection mechanisms. Hence this study helps to understand the various proteins that can be targeted for further development in drug discovery and also in the prevention of this disease.Keywords
SS Negative sense RNA; Genetic material; Nucleoprotein; Orthomyxoviridae; Alphainfluenzavirus; Homosapiens; Interaction; Infection mechanism.
Introduction
Influenza A virus (IAV) belongs to the family Orthomyxoviridae and it is the only species occurring in the genus Alphainfluenzavirus [1]. The IAV genome was ss negative sense RNA [2] and it is the most diverse and epidemically effective pathogen which causes severe respiratory disease in humans and various zoonotic infections in birds [3]. Influenza viruses circulating in human populations infect millions of people annually which causes highly expensive health consequences [4]. In the 20th century, many influenza pandemics occurred in 1918, 1957, and 1968 which merely destroyed most of the human population [5]. The first case of human infection with influenza A virus was reported in Hong Kong in 1997. Since then the virus has become a threat to humans causing 2 50,000 - 5, 00, 000 causalities worldwide [6]. There are two common mechanisms involved to acquire high pathogenicity in humans. The first mechanism involves the acquisition of adaptive mutations and genetic assortment and the other virulence mechanism was multiple viral accessory proteins which are encoded on a single gene segment [7]. The IAV genome was comprised of eight negative-sense RNA segments where each segment encodes with trimeric viral polymerases (PB1, PB2, and PA) and nucleoproteins (NP) to form viral ribonucleoprotein [8].
This is a pathogenic agent of an acute respiratory tract infection suffered by 5-20% of the world population. Although this virus is sporadic it can cause because of its strong capability to transmit from one person to another person easily [9-11]. The natural habitat of the influenza A virus is wild water flow and domestic poultry. There are various methods for treating the influenza A virus, M2 ion inhibitors, and neuraminidase inhibitors are the two major anti-influenza infection medicines [12]. Although the modern generation treatment concentrates on vaccination and usage of drugs, both treatments have some limitations because the IAV viral strains have a high mutating ability and generate resistivity against antiviral agents. Here the vaccines also required a considerable amount of time to develop to match against viral strains [5]. The viral life cycle was mainly dependent on conquering the host cell’s biological processes to facilitate its growth. IAV requires a suitable recognition site for its replication in mammalian cells. Influenza infection activates various immune responses like cytokine induction and apoptosis [6]. Thus various experimental studies were conducted on IAV strains to classify their hidden capabilities. Basing on the study of virus-host protein interactions, scientists could figure out essential factors affecting viral infections, and those factors were targeted for drug therapies[7,13]. The current aim of this study is to construct a protein interaction network of IAV with humans (host) to identify the proteins involved in the viral infection mechanism.
Materials and Methods
4.1 Data Collection
The collection of data was done using the String virus database (string-db.org), this database was a pre-computerized database that gives a better understanding of protein-protein interactions of viruses and hosts. This database especially codes for virus-virus and virus-host interactions and shows results based on experimental and text mining channels to provide combined probabilities of interactions of virus and host proteins. This database consists of 1, 17,425 interactions between 239 viruses and 319 hosts. The results were differentiated by confidence scores which indicate interactions between nodes that are connected by multiple paths and were of four categories of scores i.e. highest confidence score (0.9-1), high confidence score (0.7-0.8), medium confidence score (0.4-0.6), low confidence score (up to 0.1) [14].
4.2 Network Construction and Analysis
4.2.1 Network analysis
Cytoscape (version: 3.7.1) is public source software for integrating bio-molecular interaction networks. It is powerful when used in conjunction with large databases of proteinprotein, protein-DNA, and genetic interactions increasingly available for humans and model organisms. This software core provides basic functionality to layout and the network to visually integrate the network with expression profiles, phenotypes, and other molecular states and to link the network to databases of functional annotations. Cytoscape was provided with the various plug-in which enhances our ability to understand the network interactions. This was used to construct and visualize the IAVhuman protein-protein interaction network [15].
4.3 Topological Properties
Recently due to a rapid increase in experimental techniques and various computational methods which produce a bulk amount of interaction data, a versatile Cytoscape plug-in called network analyzer was introduced to analyze the organization and structure of complex networks. This tool analyses and produces various statistical results including numbers of nodes, edges, connecting components, the diameter of the network, density, heterogeneity, radius, average clustering coefficient, path length, neighborhood connectivities, distribution of node degrees, and betweenness centrality [16].
4.4 Cluster analysis
Cytoscape is comprised of a plug-in called MCODE (Multicontrast Delayed Enchantment) which is used to perform cluster analysis. This is used to identify the densely connected nodes from the highly-dense network. This method is based on vertex weighting by local neighborhood density and outward transversal from locally dense seed protein to isolate dense regions according to given parameters [16,17]. This helped separate biological modules or related sets of nodes. There the analysis of the network can be achieved by setting various parameters like node score cutoff, haircut, fluff, K-core, and max depth from seed [18].
4.5 Gene enrichment analysis
Gene enrichment analysis is done using a public source database called Panther GO database. Panther GO is a resource of protein analysis done through evolutionary relationships and functional classification of those genes from all organisms. In this source various properties like prokaryotic and plant genomics to phylogenetic gene trees and expanding the representation of gene evolution in these lineages and also for evolutionary classifications. Here we can analyze about 900 genomes using updated statistical tests with false discovery rate corrections per multiple testing. In this network, the clustered proteins were analyzed for proteins involved in various biological processes [19].
Results
5.1 Protein-protein Network Analysis of IAV with Homosapiens
The data regarding the protein interaction network of IAV with humans were retrieved from a public source database called STRING VIRUS database. The interaction was taken under certain conditions which include a minimum required interaction score with medium confidence of (0.400), a maximum number of interactors to be shown in the first shell is 200 and the maximum number of interactors to be shown in the second shell is 100. By reaching the required conditions a protein interaction network of 5277 interactions and 311 nodes (proteins) was resulted and was retrieved in a TSV format. This resulted network was reconstructed using public source software called Cytoscape.
5.2 Cluster analysis
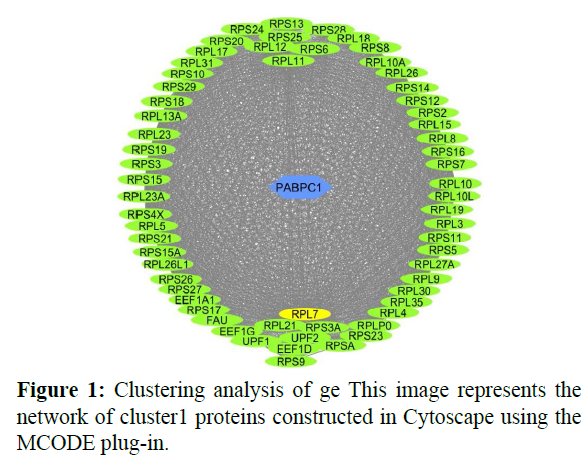
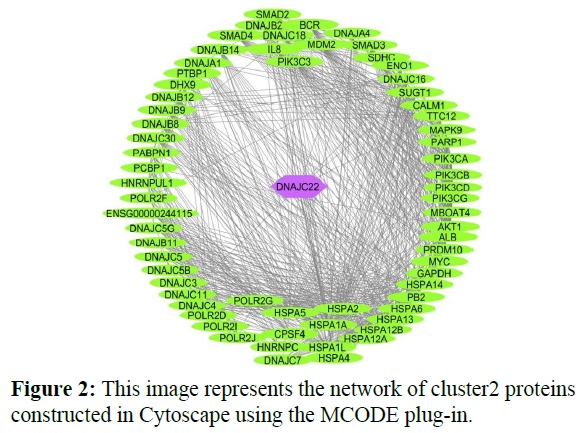
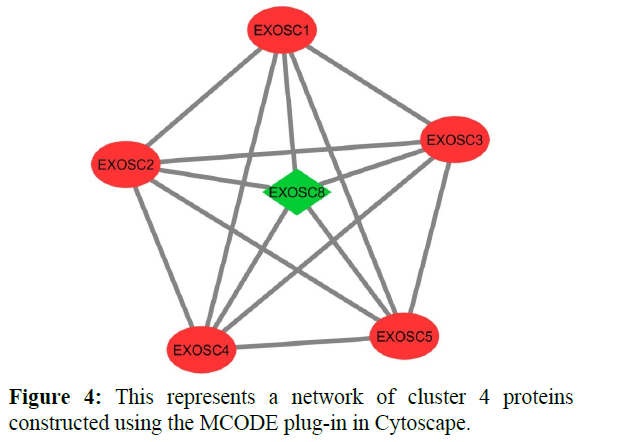
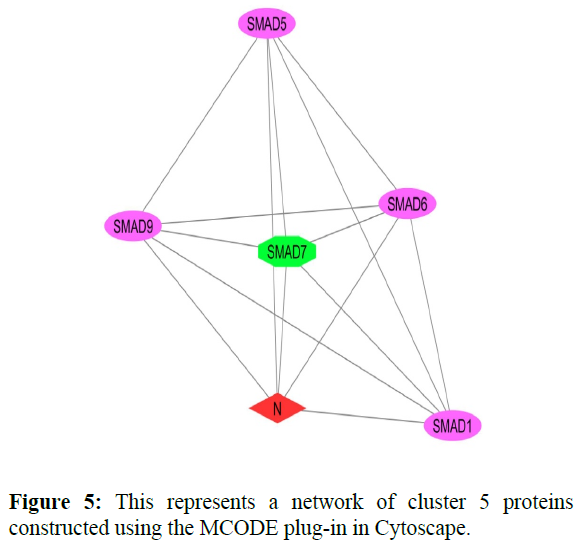
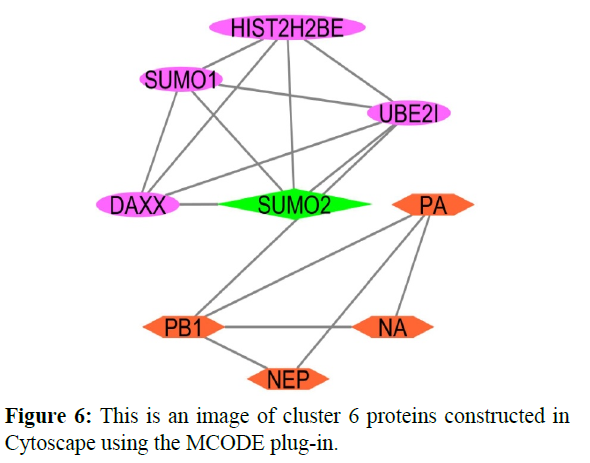
The reconstructed network was analyzed by using a plug-in called MCODE which resulted in six highly interacting clusters (C1, C2, C3, C4, C5, and C6) with a total of 179 proteins and were analyzed respectively. The first cluster has the highest interaction score of 62.419 and clustering coefficient of 0.496 and this network is comprised of 63 nodes and 1935 interactions, with a seed protein PABPC1 as shown in Figure 1. Cluster 2 has the second-highest interaction score of 20.091 and clustering coefficient of 0.413 with 67 nodes and 663 interactions and a seed protein DNAJC22 as shown in Figure 2. Cluster 2 was succeeded by cluster 3 with an interaction score of 17.455 and a clustering coefficient of 0.426 this cluster comprised 34 nodes and 288 interactions with no seed protein as shown in Figure 3. Cluster 4, cluster 5, and cluster 6 has the least interaction scores 6.0, 6.0, and 4.0, and clustering coefficients of these clusters were 0.5, 0.5, and 0.422 respectively, where clusters 4 and 5 has 6 nodes and 15 interactions each with seed proteins EXOSC8 and SMAD7 and cluster 6 has 9 nodes and 16 interactions with a seed protein SUMO2. These clusters are represented in Figure: 4, Figure: 5, and Figure: 6 respectively.
5.3 Gene enrichment analysis
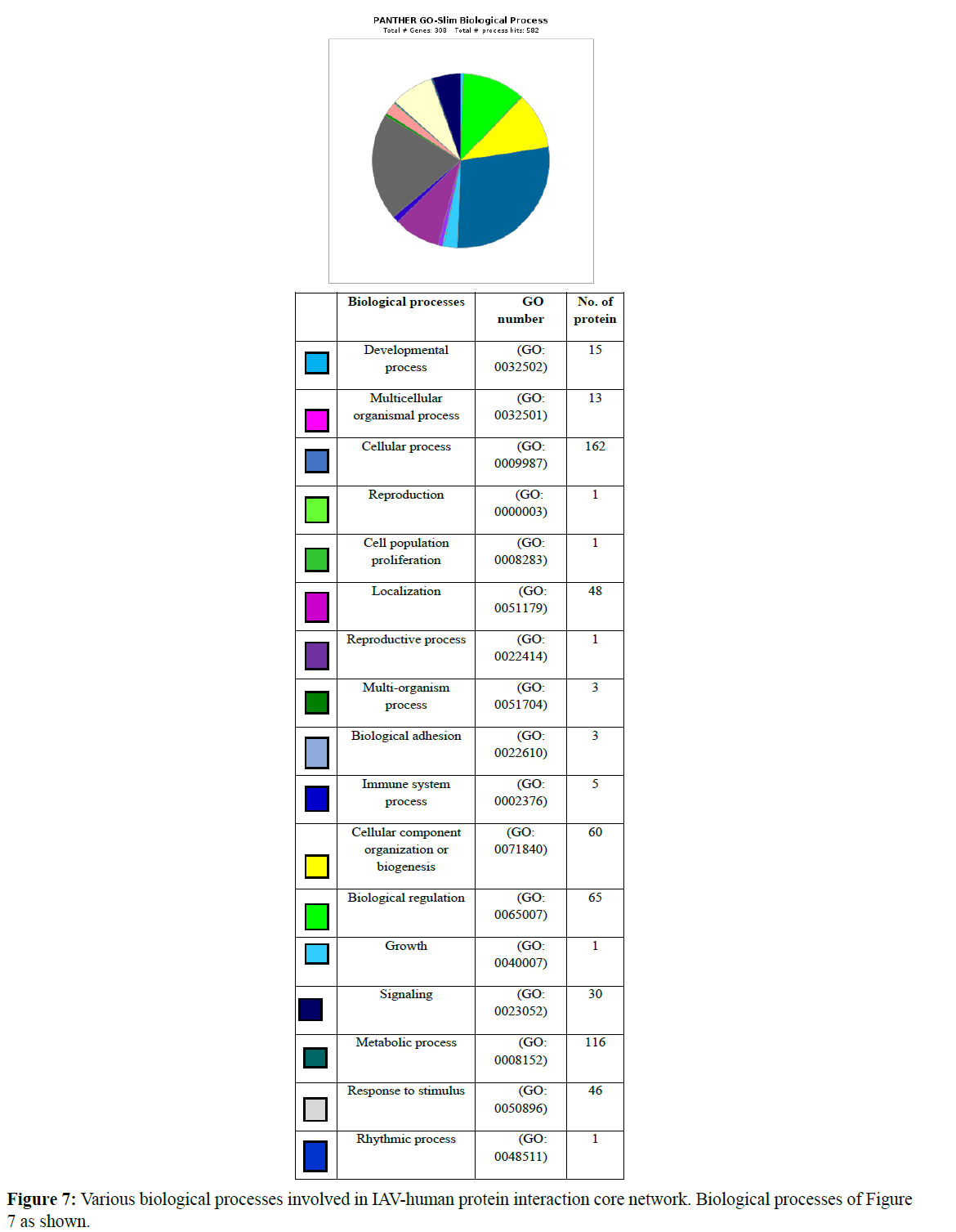
Gene ontology-based analysis is done for the resulted clustered proteins to identify their various biological processes by using the panther GO database. Here the interaction network of IAVhuman proteins was analyzed where 132 proteins were identified for cellular processes (GO: 0009987). Afterward, 179 proteins were isolated from clusters and analyzed for their respective GO term biological process and GO term cellular processes (GO:0009987) where about 110 proteins were identified which perform various cellular processes and these proteins were further analyzed for various cellular functions involved in various cellular processes concerning their clusters. Various biological processes involved in the IAV-human protein interaction core network are represented in a graphical manner Figure 7 and the number of proteins involved in each biological process was also represented.
Cluster 1 was subjected to gene enrichment analysis in which we identified 41 proteins functioning for various cellular processes. In cluster 1 „RPS3“ protein involves in cellular functions like cell communication (GO: 0007254), cell death (GO: 0008219), cellular response to stimuli (GO: 0016043), and signal transduction (GO: 0007165). RPS3 is a 40S ribosomal protein that has endonuclease activity and plays an important role in repairing damaged DNA. This displays a high binding affinity for 7, 8-dihydro-8- oxoguanine (8-oxoG) and this also stimulates the cleavage of the phosphodiester backbone by APEX1.RPS3 also helps in viral transcription in its biological processes. About 16 proteins are involved in the cellular component organization (GO: 0016043) and in the cellular metabolic process (GO: 0044237) 31 proteins were involved.
As the above cluster, cluster 2 was also analyzed for gene enrichment analysis where 32 proteins were isolated which functions for various cellular processes like cell cycle process (GO: 0007154), cell cycle (GO: 007049), cellular development process (GO: 0048869), a process utilizing autophagic mechanism (GO: 0061919), vesicle targeting (GO: 0066903) only one protein is involved. 10 proteins are involved in cell communication (GO: 0007154), 7 proteins are involved in the cellular component organization (GO: 0016043), 22 proteins are involved in the cellular metabolic process (GO: 0044237), 21 proteins are involved in cellular response stimuli (GO: 0051716), 3 proteins are involved in the microtubule-based process (GO: 0007017), 6 proteins are involved in the movement of cells or subcellular components (GO: 0006928), 11 proteins are involved in Protein folding (GO:0006457), 10 proteins are involved in Signal transduction (GO: 0007165). Here in this cluster proteins like “AKT1” a Proline-rich AKT1 substrate 1’ is a Subunit of mTORC1, which regulates cell growth and survival in response to nutrient and hormonal signals. mTORC1 is activated in response to growth factors or amino acids. Growth factor-stimulated mTORC1 activation involves AKT1-mediated phosphorylation of TSC1-TSC2, which leads to the activation of the RHEB GTPase that potently activates the protein kinase activity of mTORC1. “POLR2D” is a DNA-dependent RNA polymerase and a subunit of ‘RPB4’ catalyzes the transcription of DNA into RNA using the four ribonucleoside triphosphates as substrates. This is a component of RNA polymerase II which synthesizes mRNA precursors and many functional non-coding RNAs. “POLR2G” is a subunit of ‘RPB7’ and has the functional similarity as “POLR2D”. These three proteins are involved majorly in cellular functions and are responsible for viral transcription. “DNAJB12” is ‘DnaJ homolog subfamily B member 12’ it acts as a co-chaperone with HSPA8/Hsc70 and is required to promote protein folding and trafficking, prevent aggregation of client proteins, and promote unfolded proteins to endoplasmic reticulum-associated degradation (ERAD) pathway. It acts by determining HSPA8/ Hsc70’s ATPase and polypeptide-binding activities. “DNAJB14” a ‘DnaJ homolog subfamily B member 14’ this protein also has similar functions as DNAJB12 and both of these are involved in viral processes. Other proteins like CALM1, HSPA1A, and PPIA are involved in viral transcription, viral receptor activity, and viral lifecycle respectively. IL8 is an interleukin that regulates the ss viral RNA through dsDNA.
Cluster 3 was also undergone through gene enrichment analysis in which 27 proteins were identified involving various cellular processes. Approximately 4 proteins are involved in cell communication (GO: 0007154) and Response to stimuli (GO: 005176), 2 proteins are involved in Cellular component organization (GO: 0016043), 19 proteins are involved in Cellular metabolic processes (GO: 0044237), 5 proteins are involved in Protein folding (GO: 0006457) and 3 proteins are involved in Signal transduction (GO: 0007165). In this cluster proteins like “PSMB1, PSMB2, PSMB3, PSMB4, PSMB5, PSMB6, and PSMB7” are Proteasome subunit beta type where they are differentiated by their beta types. They are Components of the 20S core proteasome complex involved in the proteolytic degradation of most intracellular proteins. This complex plays numerous essential roles within the cell by associating with different regulatory particles. These proteins involved in interleukin-1 signaling pathways and also involved in viral processes. Other proteins like PSMA4, PSMA3, and HSP90AA1 are also involved in viral processes.
Finally clusters 4 and 6 were analyzed where 5 proteins were identified for each cluster respectively which is involved in various cellular processes. In cluster 4 five proteins are involved in the Cellular metabolic process (GO: 0044237) those are “EXOSC8, EXOSC4, EXOSC5, EXOSC3, EXOSC2”. These proteins are exosome complexes that have 3’->5’ exoribonuclease activity and participate in a multitude of cellular RNA processing and degradation events. In the nucleus, the RNA exosome complex is involved in the proper maturation of stable RNA species such as rRNA, snRNA, and snoRNA, in the elimination of RNA processing by-products and non-codingpervasive’ transcripts, such as antisense RNA species and promoter-upstream transcripts (PROMPTs), and mRNAs with processing defects, thereby limiting or excluding their export to the cytoplasm and these proteins are helpful in defensive response against viral antigens. Cluster 6 includes two types of cellular processes like Cellular component organization (GO: 0016043) and Cellular metabolic process (GO: 0044237) where 2 and 4 number proteins involved in respective functions [20-22].
The 110 proteins which have been identified for involving in various cellular processes were further analyzed to determine the proteins which are responsible for the IAV infection mechanism through the Uniprot protein database which is a public server that allows us to retrieve any information that has been dumped through various experiments done by scientists worldwide. Through analysis of those 110 proteins, we identified 674 proteins that play a major role in the IAV infection mechanism in humans. These 64 proteins can be used as targets for the discovery of various drugs which can be used for the treatment of the Influenza A virus. The 64 proteins identified are represented in Table 1.
| Protein ID | Biological Functions |
|---|---|
| RPS9,RPS15,RPL9,PSA,RPS21, RPS7,RPS10,RPS28,RPS16, RPL13A,RPL23A,RPS4X,RPL17, RPL11,RPS5,RPL31,RPS14,RPS3, RPL35,RPS8,RPL15,RPS19,RPS27, RPL10,RPL26,RPL3,RPL12,RPL17, RPL10A,RPLP0,RPS23,RPL5, RPL26L1,RPS2,AKT1,POLR2G,POLR2D. |
Viral Transcription |
| UPF1, DNAJB14, DNAJB12, CALM1, HSP90AA1, PSMA4, PSMA3, PSMB1, PSMB6, PSMB5, PSMB3, PSMB4, PSMB7, PSMB2, UBE2I, SUMO1, SUMO2, HIST2H2BE. | Viral Processes |
| IL8 | REGULATION OF ss VIRAL RNA THROUGH dsDNA |
| HSPA1A | Viral Receptor Activity |
| DNAJC3, EXOSC2, EXOSC3, EXOSC4, EXOSC5, EXOSC8. | Defensive Responses to Virus |
| PPIA | Viral Life Cycle |
| PA, PB1, NA, N | Viral Proteins |
Table 1: Various proteins involved in IAV infection and mechanism in humans.
Discussion
Influenza A virus has a variety of strains which has high transmissibility and high pathogenesis. In China, an epidemic of a novel assortment of Influenza A virus has broken out in 2013 causing a threatening infection to humans [15,23]. Scientists have discovered a vaccine and some immunization methods for the prevention and control of influenza infection but, those efforts must be updated from time to time as the IAV was a highly mutating pathogen and thus becomes resistant to the drugs discovered. The vaccines that are discovered so far were not effective on the persons who are above 65 years because of the aging factor the innate and adaptive immune responses gradually deteriorate which leads to a decrease in the ability to respond to an infection and immunization. The elderly people experience vaccine-induced immunogenicity of only 30-40%. Further vaccination may cause dysregulation of various components, deterioration of some functions, and may also cause autoimmune diseases [24]. Therefore scientists are thriving for the discovery of new drugs and therapies for the treatment of IAV infection.
The present modern antiviral drugs against IAV infection mostly target the M2 channel and neuraminidases [14,25-30].IAV requires a suitable recognition site for mammalian for its replication [6]. The IAV initiates the infection by using HA molecules present on the envelope of the viral protein. The HA receptor-binding site attaches the virus to the surface glycoconjugates that contain terminal SA residues [31,32]. The virus searches for the proper sialylated receptor by using the sialidase function of NA to remove sialic acid (SA) and to free the non-productive HA associations. The HA molecules from human IAV’s have higher specificity for receptors with α-2, 6-linked SA’s. Several studies have shown that matching HA receptor binding preferences with the SA linkages in a particular host is not essential for infection, but is more critical for transmission. Thus the IAV shows cell tropism in airways or may use more than one receptor [33-35]. After attachment, virions enter the endosomal pathway [36]. Basing on this mechanism the important IAVinteracting host proteins are being identified and used for drug targets. In previous studies, scientists identified some of the most interacting host proteins with IAV they are namely LNX2, MEOX2, TFCP2, PRKRA, and DVL2 [26].
Watanabe et al. [27] proposed the idea in which the virus-host interactions can be taken as the basement of the development of new antiviral drugs targeting host cellular factors. In this experiment, we integrate the protein interaction data by different computational methods and various databases to obtain highly IAV associated host proteins to provide information for the development of new drugs. During past decades the number of clinically valid drug targets was 324 [28], this is because the development of a new drug takes a minimum of 12-15 years with a cost of $1 billion which is a high amount to expend [29].
Here we used Cytoscape as a platform and its important plug-ins and other tools to explore the IAV-human protein interaction and found 311 host proteins that are associated with IAV, among them about 64 proteins are responsible for various IAV infection mechanisms and can be taken as potential drug targets for the cure of IAV infection. From the identified proteins 37 of them involves viral transcription, 18 proteins involves in viral processes, one protein is responsible for the regulation of viral RNA through dsDNA, 6 proteins are responsible for defensive responses to viruses, and one protein for each is responsible for both viral receptor activity and viral life cycle respectively. PA, NA, PB1, N are the viral proteins responsible for causing infection. PA is a Polymerase acidic protein in Influenza A virus which involves in biological processes like viral budding from the plasma membrane, viral genome maturation, viral genome packaging, viral RNA genome replication, viral transcription, virion attachment to host cell and also plays an essential role in viral RNA transcription and replication by forming the heterotrimeric polymerase complex together with PB1 and PB2 subunits. The complex transcribes viral mRNAs by using a unique mechanism called cap-snatching . It consists of the hijacking and cleavage of host-capped pre-mRNAs. These short-capped RNAs are then used as primers for viral mRNAs. NA is a Neuraminidase protein which involves in a biological process like viral budding from the plasma membrane and catalyzes the removal of terminal sialic acid residues from viral and cellular glycoconjugates. This helps in Cleaving the terminal sialic acid on the glycosylated HA during viral budding to facilitate its release. Additionally, it also helps the virus to spread through the circulation by further removing sialic acids from the cell surface. These cleavages prevent selfaggregation and ensure the efficient spread of the progeny virus from cell to cell. Otherwise, infection would be limited to one round of replication. NA does not play an important role in viral entry, replication, and assembly [37]. PB1 is RNA-dependent RNA polymerase that is responsible for the replication and transcription of virus RNA segments. The transcription of viral mRNAs occurs by a unique mechanism called cap-snatching [38] and also involves biological processes like negative-stranded viral RNA replication, suppression by virus of host RNA polymerase II activity, DNA-templated transcription, and viral transcription.
The proteins which were identified involves various pathways during viral infection. These proteins involved in various signaling pathways where two proteins namely AKT1 and HSPA1A involves in Apoptosis signaling pathway (P00006) and some proteins also involve in various immune response pathways in which AKT1 and CALM1 involve in T cell activation (P00053) and B cell activation (P00010) [39-41]. Two proteins namely IL8 and AKT1 involve in Inflammation mediated by chemokine and cytokine signaling pathway (P00031) [42] and Interleukin signaling pathway (P00036). EXOSC2, EXOSC3, EXOSC4, EXOSC5 involves in the RNA degradation pathway [43]; UPF1, SUMO2, UBE2I, SUMO1 involves in the RNA transport mechanism [44]. PSMA3, PSMA4 PSMB1, PSMB2, PSMB4, PSMB5, PSMB6, PSMB7 involves in the proteasome pathway [45] and RPL35 RPL13A, RPSA, RPL10A, RPL26L1, RPL3, RPL5, RPL9, RPL10, RPL11, RPL12, RPL15, RPL17, RPL23A, RPL26, RPL31, RPLP0, RPS2, RPS3, RPS4X, RPS5, RPS7, RPS8, RPS10, RPS14, RPS15, RPS16, RPS19, RPS21, RPS23, RPS27, RPS28 involves in ribosomal synthesis pathway [46]. These are the pathways that are occurring during IAV infection and play an important role in the IAV infection mechanism.
The data we structured in this study was taken from a vast variety of literature sources and various databases. Therefore the constructed IAV-human protein interaction network and identified proteins can be benefited from various new drugs.
Conclusion
Our work concentrates on proteins involved in the IAV infection mechanism in humans and was explained by computational work.
Influenza was caused by the influenza A virus which is a worldwide problem with seasonal and pandemic characteristics. These days’ scientists are using computational biology to investigate deep into the host factors responsible for viral infection.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Acknowledgements
The authors would like to thank Dr. Gollapalli Pavan for his help in the initial phase of research work. The author would like to thank the management of NITTE for providing the necessary facilities to carry out this project work.
REFERENCES
- Gutiérrez RA, Naughtin MJ, Horm SV, San S, Buchy P. A (H5N1) Virus Evolution in South East Asia. Viruses. 2009; 1(3):335-361.
- Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008; 26 Suppl 4(Suppl 4): D49-D53.
- Tscherne DM, García-Sastre A. Virulence determinants of pandemic influenza viruses. J Clin Invest. 2011; 121(1):6-13.
- Zhao M, Wang L, Li S. Influenza A Virus-Host Protein Interactions Control Viral Pathogenesis. Int J Mol Sci. 2017; 18(8):1673.
- Bright RA, Medina MJ, Xu X, et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005; 366(9492):1175-1181.
- Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010; 7(6):427-439.
- Gonzalez MW, Kann MG. Chapter 4: Protein interactions and disease. PLoS Comput Biol. 2012; 8(12):e1002819.
- Cook HV, Doncheva NT, Szklarczyk D, von Mering C, Jensen LJ. Viruses. STRING: A Virus-Host Protein-Protein Interaction Database. Viruses. 2018; 10(10):519.
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13(11):2498-2504.
- Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics.2008; (2):282-284.
- Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape3. Curr Protoc Bioinformatics. 2014; 47:8.13.1-8.13.24.
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim, and improvements in enrichment analysis tools. Nucleic Acids Res. 2019; 47(D1): D419-D426.
- Farooq QUA, Shaukat Z, Aiman S, Zhou T, Li C. A systems biology-driven approach to construct a comprehensive protein interaction network of influenza A virus with its host. BMC Infect Dis. 2020; 20(1):480.
- Hu Y, Sneyd H, Dekant R, Wang J. Influenza A Virus Nucleoprotein: A Highly Conserved Multi-Functional Viral Protein as a Hot Antiviral Drug Target. Curr Top Med Chem. 2017; 17(20):2271-2285.
- Vasin AV, Temkina OA, Egorov VV, Klotchenko SA, Plotnikova MA, Kiselev OI. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res. 2014; 185:53-63.
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003; 4:2.
- Forst CV, Zhou B, Wang M, et al. Integrative gene network analysis identifies key signatures, intrinsic networks, and host factors for influenza virus A infections. NPJ Syst Biol Appl. 2017; 3:35.
- Manzoor R, Igarashi M, and Takada A. Influenza A Virus M2 Protein: Roles from Ingress to Egress. Int J Mol Sci. 2017; 18(12):2649.
- Hu J, Ma C, Liu X. PA-X: a key regulator of influenza A virus pathogenicity and host immune responses. Med Microbiol Immunol. 2018; 207(5-6):255-269.
- Qu XY, Li QJ, Zhang HM, et al. Protective effects of phillyrin against influenza A virus in vivo. Arch PharmRes. 2016; 39(7):998-1005.
- Kuo SM, Chen CJ, Chang SC, et al. Inhibition of Avian Influenza A Virus Replication in Human Cells by Host Restriction Factor TUFM Is Correlated with Autophagy. MBio. 2017; 8(3):e00481-17.
- Molinari NA, Ortega-Sanchez IR, Messonnier ML, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007; 25(27):5086-5096.
- Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013; 368(20):1888-1897.
- Smetana J, Chlibek R, Shaw J, Spline M, Prymula R. Influenza vaccination in the elderly. Hum Vaccin Immunother. 2018; 14(3):540-549.
- Rungrotmongkol T, Intharathep P, Malaise M, et al. Susceptibility of antiviral drugs against 2009 influenza A (H1N1) virus. Biochem Biophys ResCommun. 2009; 385(3):390-394.
- Farooq QUA, Shaukat Z, Aiman S, Zhou T, Li C. A systems biology-driven approach to construct a comprehensive protein interaction network of influenza A virus with its host. BMC Infect Dis. 2020; 20(1):480.
- Watanabe T, Kawaoka Y. Influenza virus-host interactomes as a basis for antiviral drug development. Curr Opin Virol. 2015; 14:71-78.
- Li Q, Lai L. Prediction of potential drug targets based on simple sequence properties. BMC Bioinformatics. 2007; 8:353.
- Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. Br J Pharmacol. 2011; 162(6):1239-1249.
- Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010; 7(6):440-51.
- Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. 2010; 285(37):28403-9.
- Hamilton BS, Whittaker GR, Daniel S. Influenza virus-mediated membrane fusion: determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses. 2012; 4(7):1144-68.
- Dou D, Revol R, Östbye H, Wang H, Daniels R. Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement. Front Immunol. 2018; 9:1581.
- Linster M, van Boheemen S, de Graaf M, Schrauwen EJA, Lexmond P, Mänz B, Bestebroer TM, Baumann J, van Riel D, Rimmelzwaan GF, Osterhaus ADME, Matrosovich M, Fouchier RAM, Herfst S. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell. 2014; 157(2):329-339.
- Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solórzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, García-Sastre A. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007; 315(5812):655-9.
- Rust MJ, Lakadamyali M, Zhang F, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 2004; 11(6):567-73.
- Liu C, Eichelberger MC, Compans RW, Air GM. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol. 1995; 69(2):1099-106.
- Li L, Dai H, Nguyen AP, Hai R, Gu W. Influenza A virus utilizes noncanonical cap-snatching to diversify its mRNA/ncRNA. RNA. 2020; 26(9):1170-1183.
- Othumpangat S, Beezhold DH, Noti JD. Influenza virus infection modulates the death receptor pathway during the early stages of infection in human bronchial epithelial cells. Physiol Genomics. 2018; 50(9):770-779.
- Chen X, Liu S, Goraya MU, Maarouf M, Huang S, Chen JL. Host Immune Response to Influenza A Virus Infection. Front Immunol. 2018; 9:320.
- Lin C, Zimmer SG, Lu Z, Holland RE Jr, Dong Q, Chambers TM. The involvement of a stress-activated pathway in equine influenza virus-mediated apoptosis. Virology. 2001; 287(1):202-13.
- Format C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010; 115(11):2167-76.
- May JP, Yuan X, Sawicki E, Simon AE. RNA virus evasion of nonsense-mediated decay. PLoS Pathog. 2018; 14(11):e1007459.
- Hutchinson EC, Fodor E. Transport of the influenza virus genome from the nucleus to nucleus. Viruses. 2013; 5(10):2424-46.
- Gao G, Luo H. The ubiquitin-proteasome pathway in viral infections. Can J Physiol Pharmacol. 2006; 84(1):5-14.
- Li S. Regulation of Ribosomal Proteins on Viral Infection. Cells. 2019; 8(5):508.