Using Deep Learning for Automatic Icd-10 Classification from Free-Text Data
2 Health Management Center, National Taiwan University Hospital, Taipei, Taiwan, Email: gkclu@mail.ncku.edu.tw
3 Department of Computer Science and Information Engineering, National Taiwan University, Taipei, Taiwan, Email: yvchen@andrew.cmu.edu
4 Medical Information Management Offices, NTUH, Taipei, Taiwan
5 Information Technology Office, NTUH, Taipei, Taiwan, Email: leechung@usc.edu
6 Department of Electrical Engineering, Tunghai University, Taichung, Taiwan, Email: yufang-chung@uiowa.edu
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact submissions@ejbi.org
Abstract
Background: Classifying diseases into ICD codes has mainly relied on human reading a large amount of written materials, such as discharge diagnoses, chief complaints, medical history, and operation records as the basis for classification. Coding is both laborious and time consuming because a disease coder with professional abilities takes about 20 minutes per case in average. Therefore, an automatic code classification system can significantly reduce the human effort. Objectives: This paper aims at constructing a machine learning model for ICD-10 coding, where the model is to automatically determine the corresponding diagnosis codes solely based on free-text medical notes. Methods: In this paper, we apply Natural Language Processing (NLP) and Recurrent Neural Network (RNN) architecture to classify ICD-10 codes from natural language texts with supervised learning. Results: In the experiments on large hospital data, our predicting result can reach F1-score of 0.62 on ICD-10-CM code. Conclusion: The developed model can significantly reduce manpower in coding time compared with a professional coder.
Keywords
ICD-10; Machine learning; Neural network; Natural language processing; Text classification
Introduction
Most medical researchers store data in a structured manner, such as birthday, height, weight, and drug concentration in blood or blood oxygen level of the patients. Typically, structured data are efficient for data analysis because it is usually organized in a relational database, which is easy to be retrieved and analyzed. Therefore, most of the traditional algorithms worked well with structured data. In contrast, unstructured data may be ambiguous and irregular, which contains a paragraph about a patient’s history of diseases written in English or Chinese. People with background knowledge can realize and extract the features from these free-text data; however, this task is challenging for machines.
Such task of information extraction from free-text data has been widely studied in natural language processing (NLP). NLP is a research area of research and application that makes computer systems understand and manipulate natural language text or speech to perform desired tasks [1]. The models in NLP have attempted at identifying critical elements in documents, summarizing information in documents into abstracts, or translating texts to a different language.
Considering achievements in the NLP area, applying NLP techniques is beneficial to understand the semantics in the unstructured data from the medical domain.
In the past, many medical researchers used NLP to solve their problems, such as summarizing a long paragraph like clinical notes or academic journal articles, by identifying keywords or concepts in the free-form texts [2].
Recently, NLP models have become more advanced in extracting meanings from unstructured healthcare data, and more medical data can be used for model training [3]. Therefore, computers can gradually take over more routine jobs, which could only be done by humans in the past.
Electronic health records (EHR) are the collection of patients and electronically-stored health information of population in a digital format. By the digital format, medical data can accurately provide the latest patients information and also allows medical researchers to extract the required information in hospitals to be shared across different medical centers to speed up the progress in academic research.
In general, EHRs contain multi-types of data, including medical history, medication and allergies, immunization status, laboratory test results, radiology images, vital signs, and personal statistics like age, weight, and billing information. The information of patients’ diseases can be extracted from the types of data and expressed with The International Statistical Classification of Diseases and Related Health Problems 10th Revision (ICD-10) code, which defines the classifying standard of diagnosis for establishing the standard expression of diagnosis for international clinical research, evaluating health care quality, and, the most important, applying for health insurance subsidies.
ICD-10 coding is a multi-class and multi-label problem, where each case may be associated with multiple codes. Besides, the ICD-10 annotations are highly unbalanced, because most frequent codes have more samples. In our data, each case contains about 1 to 20 codes from A00 to Z99 [4]. The complicated coding procedure is a time-consuming task in hospitals. Therefore, to take advantage of the capability of current NLP models, our goal is to build a model to classify text data into ICD-10 codes automatically. In previous work of deep learning and NLP related ICD code predicting task, Zhang et al. use GRU with content-based attention to predict medication prescription given the disease codes [5] and Yanshan et al. apply and make a comparison between NLP techniques such as GloVe on EHR data classification task [6].
In this paper, we focuses on applying NLP and neural networks to understand the meaning behind the unstructured data written by doctors.
Background
The International Statistical Classification of Diseases and Related Health Problems (ICD) is a medical classification list released by the World Health Organization (WHO). It contains codes for the universe of diseases, signs and symptoms, abnormal findings, complaints, social circumstances, and external causes of injuries or other related health conditions based on medical materials written by physicians such as discharge diagnosis, admission diagnosis, etc. Hence, more detailed and standardized information are provided for measuring healthcare service quality, safety and efficacy. Since the first publication in 1893, ICD was widely used in fields such as health insurance. These classified data of ICD code can be applied to the clinical management system or be an evaluation factor for the health care quality. Also, since the bureau of national health Insurance, Taiwan started to use ICD code as a reference when evaluating the amount of premium subsidies in the Diagnosis-Related Groups (DRGs) prospective payment system, ICD code has become one of the most important indexes for the hospital to apply for reimbursement.
When a patient visits a hospital for medical treatment, a series of medical data would be generated during diagnosis, such as chief complaints, history, pathology reports, discharge notes, ICU notes, etc. These medical records in each country are generally similar but written in different languages. Such free-text data contains rich information, but it is difficult to analyze by data scientists. Generally, only domain experts are able to extract the hidden message from the free-text data. In every hospital, there is a group of professional disease coders with license spending plenty time on reading discharge notes and classifying into ICD codes. Within expectation, maintaining these coders requires time and money in a certain extent. Currently, ICD codes that are used to apply for DRGs for inpatients mainly relies on professional disease coders coding case by case. However, some cases, especially outpatients, are still coded by physicians. Without professional training, cases coded by physicians tend to be incorrect and lead to cause considerable loss on benefits of hospital. As mentioned previously, approach for improving disease coding quality and reducing cost of maintaining disease coders is still a burning issue to the medical system.
There are 22 chapters in ICD-10-CM. Table 1 shows the chapter number in ICD-10-CM. ICD-10 code sets differ a lot from ICD-9 due to their fundamental changes in the structure and concepts. ICD-9-CM is composed of numeric symbols, so all ICD-9 codes are not enough to express the complicated categories of diseases. The conversion from ICD-9 to ICD-10 increases specificity to clinical diagnoses, thus creating a multitude of new codes to learn and implement. Where ICD-9-CM only has 13,000 codes, ICD-10-CM boasts 68,000. There are many alterations to the specificity and expansiveness in ICD-10 to deal with the latest diseases and procedures.
| Ch. Blocks | Title | Ch. Blocks | Title |
|---|---|---|---|
| I. A00-B99 | Certain infectious and parasitic diseases | XII. L00-L99 | Diseases of the skin and subcutaneous tissue |
| II. C00-D48 | Neoplasms | XIII. M00-M99 | Diseases of the musculoskeletal system and connective tissue |
| III. D50-D89 | Diseases of the blood and blood-forming organs and certain disorders involving the immune mechanism | XIV. N00-N99 | Diseases of the genitourinary system |
| IV. E00-E90 | Endocrine, nutritional and metabolic | XV. O00-O99 | Pregnancy, childbirth and the puerperium |
| diseases | |||
| V. F00-F99 | Mental and behavioral disorders | XVI. P00-P96 | Certain conditions originating in the |
| perinatal period | |||
| VI. G00-G99 | Diseases of the nervous system | XVII. Q00-Q99 | Congenital malformations, deformations and chromosomal abnormalities |
| VII. H00-H59 | Diseases of the eye and adnexa | XVIII. R00-R99 | Symptoms, signs and abnormal clinical and laboratory findings, not elsewhere classified |
| VIII. H60-H95 | Diseases of the ear and mastoid process | XIX. S00-T98 | Injury, poisoning and certain other |
| consequences of external causes | |||
| IX. I00-I99 | Diseases of the circulatory system | XX. V01-Y98 | External causes of morbidity and mortality |
| X. J00-J99 | Diseases of the respiratory system | XXI. Z00-Z99 | Factors influencing health status and |
| contact with health services | |||
| XI. K00-K93 | Diseases of the digestive system | XXII. U00-U99 | Codes for special purposes |
Table 1: 22 chapters in ICD-10-CM codes.
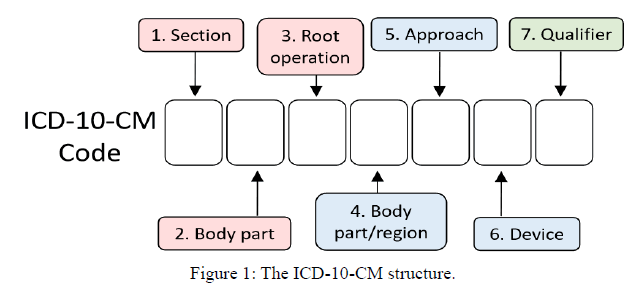
ICD-10 codes are divided into two major categories, CM and PCS, CM denotes “Clinical Modification”, while PCS denotes “Procedure Coding System”. ICD-10-CM is about the diagnosis of diseases, and its structure is illustrated in Figure 1. The first three characters of an ICD-10 code designate the category of the diagnosis. The next three characters correspond to the related etiology. The seventh character provides the extensions. Compared with ICD-9-CM which has only 3-5 characters, ICD- 10-CM has 3-7 characters respectively. Therefore, the ICD-10- CM that describes the detailed clinical information may increase the complexity of determining codes.
ICD-10-PCS has approximately 87,000 codes. Each character can be any of 34 possible values of ten digits 0-9 and 24 letters A-H, J-N, and P-Z, which may be used in each character. The letters O and I are excluded to avoid confusion with the numbers of 0 and 1.
ICD-10-PCS has approximately 87,000 codes. Each character can be any of 34 possible values of ten digits 0-9 and 24 letters A-H, J-N, and P-Z, which may be used in each character. The letters O and I are excluded to avoid confusion with the numbers of 0 and 1.
Apart from research and analyses, these coded materials can also serve as a basis for medical institutions to apply for the reimbursement of patient insurance. ICD-10 codes comprise detailed disease cases such as W59.22XA, which means “struck by a turtle”, so a large number of ICD-10 codes are rarely used that it is hard to remember all ICD-10 codes and assign them correctly. The WHO provides detailed information about ICD and a set of available materials, such as ICD-10 browser [4], online. In this website, it is obvious that ICD-10 is a huge resource that requires a lot of effort for training an expert.
Until now, classifying diseases has mainly relied on people reading amounts of written materials, such as discharge diagnoses, chief complaints, medical history, and operation records as the basis for classification. Coding is both laborious and time-consuming, considering that classifying a disease with professional abilities also takes an average of 20 minutes. Our study aims to construct an ICD-10 coding system for classifying disease information into ICD-10 codes using unitary free-text data automatically in order to retrench funds and labors in the hospital. Due to the large number of ICD-10 codes, supervised machine learning techniques are used for classifying the ICD-10 codes, instead of rule-based techniques.
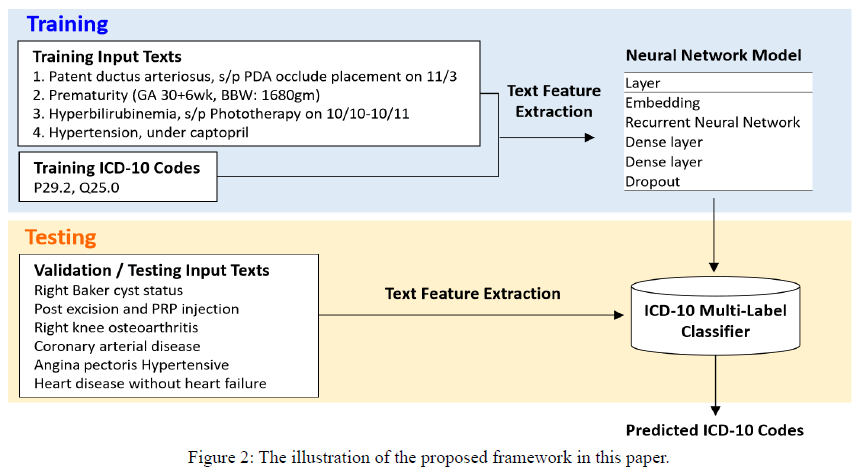
In this paper, we focus on how to build a machine learning model for an ICD-10 coding system, where the input is the free-text data describing patient’s situation and the output contains one or multiple ICD-10 codes corresponding to patient’s diseases. The proposed framework is illustrated in Figure 2. In this graph, the left part is the free-text data written by doctors. When disease classifiers are coding, they have to read a lot of free-text data like these to classify the final codes like the right part of the graph. The free-text data contains lots of information. We expect our model could learn the information behind the natural language and predict the correct codes for each patient.
Past research has already built a model for the ICD-9 system [7], but this model is built using rule-based approach. Compared with ICD-9, ICD-10 has much more codes. Building a rule-based automatic system is not an easy work. They also need to query reference tools. The whole rules of ICD-10 are so complex for humans. To solve this problem, we tend to use computers for this work. In our research, we use machine learning method to learn those rules. We try to solve this problem in mathematical approaches. Unlike the rule-based system, machine learning is closely related to computational statistics, predicting the codes due to the distribution of the data.
Materials and Methods
This section describes the collected data and then details the proposed approach.
Data Description and Preprocessing
Our data was acquired from the patients at the National Taiwan University Hospital (NTUH), where the patient data annotated with ICD-10 from January 2016 to July 2017 is used. Our data contains account IDs, chief complaints, course and treatment, history, pathology reports, physical examinations, discharge diagnoses, and transfer out of ICU diagnosis. The ground-truth ICD-10 codes are annotated by the coders in NTUH.
Most medical records were written in English, and a small part was in Chinese, where most of the Chinese words were used for recording names of hospitals in Taiwan. We, therefore, removed all of the Chinese words in our data. The null or duplicate elements, punctuation, and stop words were further removed. After those preprocessing, we could tokenize the texts and train the word2vec model for text classification. Table 2 shows the data distribution of 7 types in ICD-10 codes. The minimum number of ICD-10 class is H60 to H95 and the maximum number of ICD- 10 class is C00 to D48.
| 21 Class | Chief Complaint | Pathology Report | Physical Examination | Discharge Diagnosis | Progress | Transfer out of ICD diagnosis | History |
|---|---|---|---|---|---|---|---|
| A00-B99 | 45,349 | 10,716 | 44,999 | 45,344 | 40,541 | 4,368 | 16,964 |
| C00-D48 | 1,71,116 | 55,615 | 1,70,597 | 1,71,112 | 1,67,047 | 5,651 | 58,323 |
| D50-D89 | 25,779 | 5,828 | 25,491 | 25,779 | 23,469 | 1,928 | 8,335 |
| E00-E90 | 90,281 | 21,030 | 88,981 | 90,268 | 86,246 | 7,489 | 37,928 |
| F00-F99 | 13,465 | 1,877 | 11,722 | 13,465 | 13,011 | 747 | 4,472 |
| G00-G99 | 22,436 | 4,145 | 21,056 | 22,436 | 21,178 | 2,624 | 8,782 |
| H00-H59 | 17,230 | 1,847 | 17,040 | 17,230 | 16,922 | 525 | 8,899 |
| H60-H95 | 3,994 | 1,029 | 3,946 | 3,994 | 3,879 | 210 | 1,787 |
| I00-I99 | 1,55,384 | 33,851 | 1,52,942 | 1,55,372 | 1,48,558 | 17,725 | 68,588 |
| J00-J99 | 51,696 | 12,706 | 51,308 | 51,692 | 46,602 | 7,567 | 21,233 |
| K00-K93 | 69,602 | 21,615 | 69,084 | 69,602 | 65,918 | 4,577 | 29,639 |
| L00-L99 | 12,839 | 2,879 | 12,697 | 12,836 | 12,140 | 562 | 4,998 |
| M00-M99 | 42,279 | 7,300 | 41,472 | 42,279 | 41,097 | 1,806 | 17,584 |
| N00-N99 | 65,374 | 17,398 | 64,656 | 65,368 | 61,960 | 4,870 | 27,659 |
| O00-O99 | 13,779 | 1,180 | 13,772 | 13,779 | 13,749 | 118 | 8,269 |
| P00-P96 | 9,343 | 324 | 9,343 | 9,343 | 8,738 | 2,557 | 5,618 |
| Q00-Q99 | 12,385 | 2,735 | 12,348 | 12,385 | 11,956 | 2,365 | 6,569 |
| R00-R99 | 45,285 | 11,186 | 44,297 | 45,281 | 40,943 | 5,515 | 17,173 |
| S00-T98 | 34,050 | 5,041 | 33,628 | 34,050 | 31,867 | 3,572 | 15,308 |
| U00-U99 | 174 | 11 | 83 | 174 | 167 | 2 | 44 |
| V01-Y98 | 19,309 | 2,483 | 19,110 | 19,310 | 18,196 | 1,823 | 9,602 |
| Z00-Z99 | 1,22,025 | 17,938 | 1,21,393 | 1,22,025 | 1,18,999 | 5,108 | 48,112 |
| All data | 2,39,597 | 63,547 | 2,37,087 | 2,39,592 | 2,35,185 | 11,404 | 1,11,469 |
Table 2: ICD-10 code distribution in 7 types of data.
Other types of data have similar distribution with discharge diagnoses in 21 categories, but have different maximum length of sentences and unique words. Table 3 shows the maximum length and unique words in 7 types of documents. Longer length of sentences need a complex model, and more unique words indicate more or diverse information stored in documents.
| Free-text data | Maximum length | Mean length | Unique words |
|---|---|---|---|
| Chief complaint | 422 | 12 | 4,666 |
| Course and treatment | 624 | 119 | 21,514 |
| History | 33,141 | 312 | 30,921 |
| Pathology report | 543 | 15 | 3,023 |
| Physical examination | 641 | 181 | 9,179 |
| Discharge diagnosis | 531 | 48 | 14,858 |
| Transfer out of ICU diagnosis | 434 | 57 | 4,048 |
Table 3: Maximum length and unique words in 7 types of documents.
Feature Extraction from Discharge Notes
In order to simulate coder’s work in hospitals, our goal is to construct a model that predicts ICD-10 codes based on the given free-form texts. In our model, we first apply basic preprocessing methods via NLTK [8], and then build a neural network model for learning the features from input texts. The preprocessing procedure includes spell checking, converting into lower cases, stop words removal, tokenization, and removing infrequent words. The preprocessed data are then split to training and validation set by Scikit-Learn library.
In the neural network model, the first layer is the word embedding layer, which is the collective name for a set of language modeling and feature learning techniques in NLP where words or phrases from the vocabulary are mapped into vectors of real numbers [9]. We then encode each tokenized word into its word embedding based on word2vec and GloVe [10], considering their capability of capturing semantics and syntactic in vectors.
In our work, we obtain the best performance by using 300-dimensional embedding to represent words in the whole documents. In the word2vec model training, we use the window size parameter as 10, indicating that the maximum distance between the current and predicted word is within a sentence. The min count parameter is 5, indicating that the model ignores all words with total frequency lower than 5. The sample parameter is 0.1 which means the threshold for configuring higher-frequency words being randomly down sampled. After the training, we have the word embedding layer, which is the top layer in our neural network model to transform all of our free-text data into vector format, and our model then learns the hidden information in the documents.
In our work, we obtain the best performance by using 300-dimensional embedding’s to represent words in the whole documents. In the word2vec model training, we use the window size parameter as 1, indicating that the maximum distance between the current and predicted word is within a sentence. The min count parameter is 5, indicating that the model ignores all words with total frequency lower than 5. The sample parameter is 0.1 which means the threshold for configuring higher-frequency words being randomly down sampled. After the training, we have the word embedding layer, which is the top layer in our neural network model to transform all of our free-text data into vector format, and our model then learns the hidden information in the documents.
Deep Neural Network Model
Supervised learning [11] learns a function that maps an input to an output based on the input-output pairs. We should prepare the dataset with labeled training data. In this research, our data were labeled by the disease coders in NTU hospital from January 2016 to June 2017. Each of the free-text data had a pair of ICD-10 codes as the label. Our neural network model analyzed the input free-text data to learn a mapping function that could map the free-text data into the correct multiple ICD-10 codes. Like the concept of human learning, our model could observe the input data and correct the thought about the input data by the label, and finally understand the relationship between the input data and the output label.
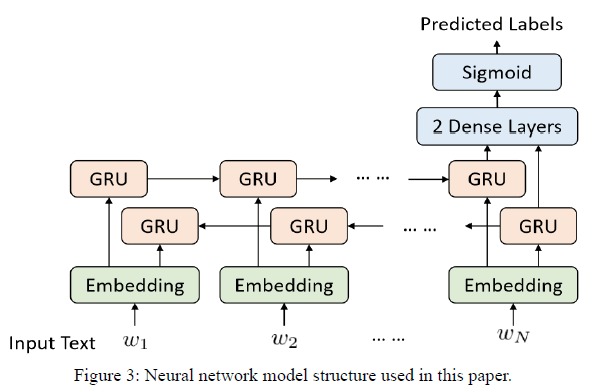
Our model structure is a four-layer neural network model which is shown in Figure 3. The first layer is the word embedding layer, which transforms the free-text input into word vectors. The second layer is a bidirectional gated recurrent unit (GRU) layer [12]. GRU is a recurrent neural network with gating mechanisms, which can solve the vanishing gradient problem that sometimes comes with the standard recurrent neural network. GRU also consumes less time for calculation than the long short-term memory (LSTM) [13]. The remaining layers are two dense layers with rectified linear unit (ReLU) and sigmoid as activation function separately, where the final dense layers should output the vector with the dimension we expect to predict. In 21 categories classification case, there are total 21 categories of ICD-10, so the final dense layer should output a 21-dimensional vector, where each dimension indicates how much probability of a code is associated with. In whole label classification case, size of output should be equal to amount of the labels, i.e. Dense layer 2 in Table 4. Table 4 shows the parameters of dropout and the four layers of our model.
| Hyperparameters | Size |
|---|---|
| Embedding layer | 300 |
| Bidirectional GRU layer | 256 |
| Dense layer 1 | 700 |
| Dense layer 2 | 14,602 |
| Dropout | 0.2 |
Table 4: Hyperparameters of whole label classification model.
Results and Discussion
To evaluate the performance our model achieves, we use F1 score as the evaluation metrics. F1-score is the harmonic mean of recall and precision. Precision and recall can evaluate the model performance with false positive and false negative. Hence, we believe that F1 score that balances both metrics is proper for our goal.
Below we investigate three experimental settings for diagnosis prediction.
ICD-10 Category Classification
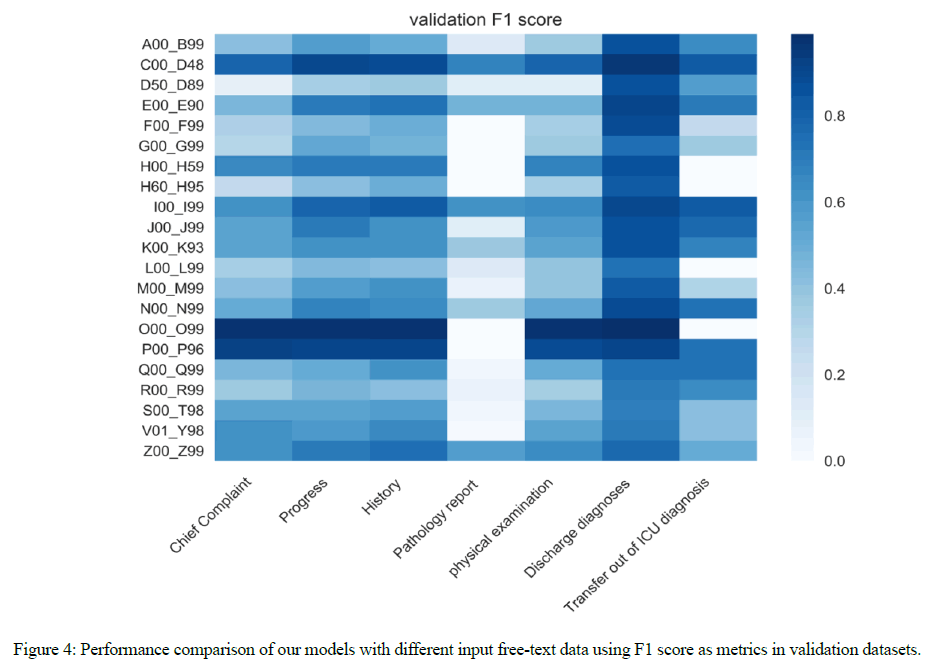
First, we category the codes based on ICD chapters. In ICD-10 CM, there are 22 blocks to subdivide codes into the format of 3 characters. Each of blocks has its title presenting the disease. For example, A00-B99 is about “Certain infectious and parasitic diseases”. Considering that U00-U99 blocks being about “Codes for special purposes” are not related to diseases, our model does not predict the ICD-10 codes between U00 to U99. Hence, our model only predicts 21 categories in ICD-10 and the validation performance is shown in Figure 4.
As Figure 4 shows, we obtain F1-score of 0.86 on the average of 21 chapters when we use discharge diagnoses as our input data. We achieve over 0.5 on F1- score in H60 to H95 and P00-P96 blocks, which only have 1,820 and 2,275 samples in our dataset.
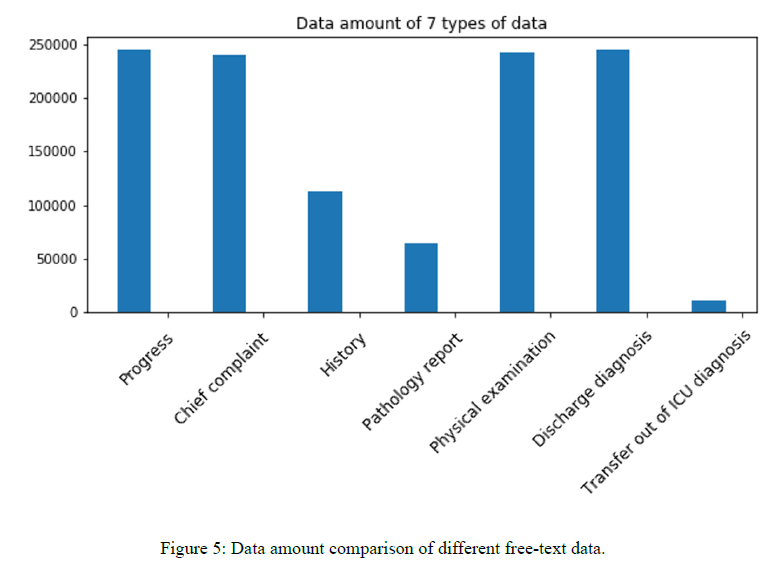
Figure 5 shows the data amount of 7 types of input data. Based on Figure 4 and Figure 5, we observe the strong correlation between performance and the data size, where the distribution implies that data amount has an effect on the training progress and the final performance. Less data records lead to worse performance in our experiments.
ICD-10 First 3 Codes Classification
The first three characters of an ICD-10 code designate diagnosis category. For example, A00 indicates “Cholera”, and A00 can be further expanded to A00.0, A00.1 or A00.9. Because better performance achieved by inputting discharge diagnoses shown in 21-category classification, we use discharge diagnoses to train the three-character classification model. There are 1,598 labels and our model has 0.715 of F1-score.
Full ICD-10 Classification
The complete ICD-10-CM code can have 3-7 characters. We have 14,602 labels in total as prediction candidates in our dataset. Similarly, we use discharge diagnoses as our input to train this model, considering better chapter classification performance and the GPU hardware bottleneck. Our model has 0.625 on F1-score when we use 300 as our embedding dimension.
Performance in Department
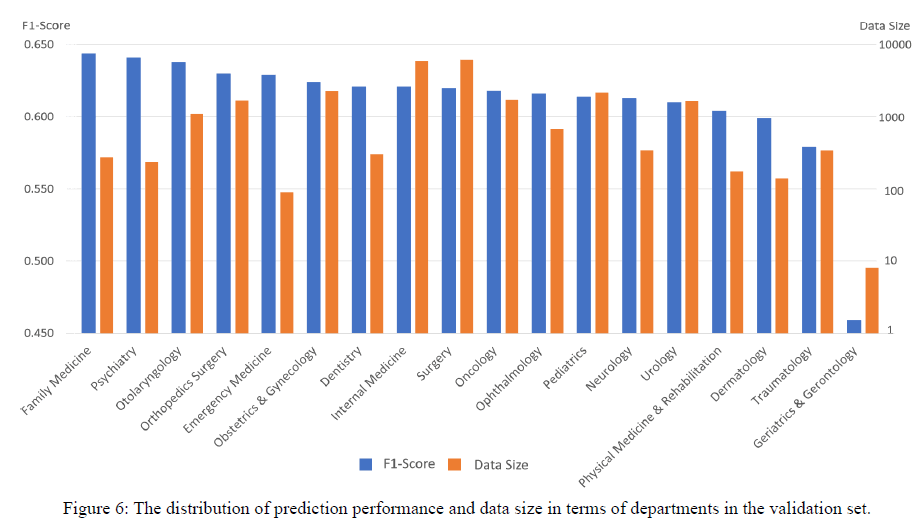
Our training and validation data covers most departments of NTUH. To further examine whether our model can generalize to different departments, we show F1-score of each department in Figure 6.
The outcome implies that data amount shows no significant effect on prediction results, and our full ICD model can achieve F1- score over 0.61 for most departments when the testing data over about 100 records, except of Department of Traumatology and Department of Dermatology. The results demonstrate that our model can generalize to different departments.
Case Discussion
In ICD chapter classification, our model achieves F1-score of 0.86, but only 0.66 in full ICD-10 prediction. In Table 5, we compare the results between the predictions from our model and professional coders’ labels.
| Prediction | Label | Free-Form Text |
|---|---|---|
| C50.911, | C50.911, C50.912, D50.9, D56.9, D63.0, F41.9, Z22.51 | 1. Advanced breast cancer, invasive carcinoma, ER (+,25%); PR (+,15%); Her2/neu (3+/3+), stage IV, under Tamoxifen (2016/11/22~), status post Herceptin + Paclitaxel (C1D1=2016/11/25) |
| C50.912, D50.9, D56.9, | 2. HBV carrier, under entecavir (since 2016/11/23~) | |
| F41.9, Z22.51 | 3. Anxiety, under control with Alprazolam | |
| 4. Microcytic anemia, suspected cancer related, but Thalassemia should be ruled out | ||
| Q25.0 | P29.2, Q25.0 | 1. Patent ductus arteriosus, s/p PDA occluder placement on 11/3 |
| 2. Prematurity (GA 30+6wk, BBW: 1680gm) | ||
| 3. Hyperbilirubinemia, s/p phototherapy on 10/10-10/11 | ||
| 4. Hypertension, under captopril | ||
| I25.10 | I10, I25.119, M17.11, M71.21 | Right Baker cyst status post excision and PRP injection Right knee osteoarthritis Coronary arterial disease Angina pectoris Hypertensive heart disease without heart failure |
Table 5: Model prediction and professional coders’ labels with the associated input texts.
In case 1, our model misses D63.0 in the prediction, which means “Anemia” in neoplastic disease. Our model does not learn from the free-text data “Microcytic anemia, suspected cancer related”. Raising the model complexity may improve the learning ability from the free-text data. In case 2, our model misses five codes of C77.2, C78.01, C78.02, C78.7, R18.8, Z51.5 and incorrectly predicts C78.00 and Z51.11. In this case, discharge diagnoses are not enough for the full ICD-10 prediction, because the model needs not only discharge diagnoses but also other data, like image reports. In case 3, P29.2 means Neonatal hypertension which occurs in babies. Our model requires additional information about this patient’s age, so only Q25.0 is correctly predicted from the free text data “patent ductus arteriosus”. In case 4, I25.119 is “Atherosclerotic heart disease of native coronary artery with unspecified angina pectoris”. This code is a combination code which describes two diagnoses in a single code. Combination is an important rule not used in ICD-9. In ICD-10, there is lot of cases using combination codes to record patients’ diagnoses. Our model cannot learn the rules from free-text data so far.
In the future work, collecting more data for training and overcoming the hardware bottleneck issue are our main targets in order to improve the model performance.
Embedding Visualization
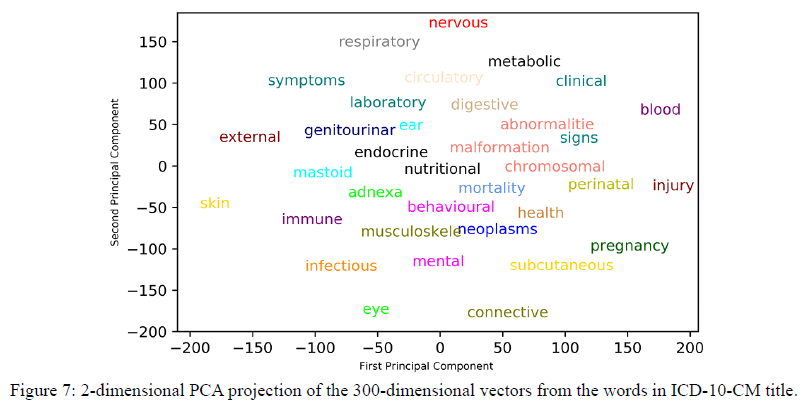
To investigate our word embedding performance, we transform our 300-dimensional word vectors into 2-dimensional vectors by the principal component analysis (PCA). We choose the words in the ICD-10-CM title in the 21 categories as shown in Figure 1.
In Figure 7, “abnormalities”, “malformation” and “chromosomal” are words in the title of the Q00-Q99 chapter. These three words are clustered obviously. We can inspect our vector performance mapping to natural language via such visualization. Even though the embedding’s learned from our data are not perfect, we believe that collecting more data can improve the vector representations.
To evaluate the performance of our word vector model, we both project our word vectors into a 2-dimensional space and calculate cosine similarity between two words to analyze the performance of word2vec. We list top 5 words similar to “cancer” in Table 6.
| Target word | Neighbors (cosine similarity) |
|---|---|
| Cancer | ivb (0.641), tumor (0.594), iva (0.585), iiic (0.576), iib (0.550) |
| Thrombosis | thrombus (0.538), dvt (0.500), thromboembolism (0.495), embolism (0.484), regrafting (0.475) |
| Stroke | cva (0.656), infarct (0.650), infarctions (0.628), tia (0.627), cardiomyopathies (0.598) |
| Allergy | hypersensitivity (0.676), eruption (0.612), interaction (0.552), anaphylaxis (0.544), bronchospasm (0.529) |
Table 6: Top 5 word similar to “cancer” through word2vec.
From the list, “ivb”, “iva”, “iiic” and “iiib” are words related to the cancer stage. The results demonstrate that the word embedding’s successfully capture the salient features from natural language in our free-text data. Unlike ICD-10 rules in other countries, each case can have at most 20 ICD-10-CM and 20 ICD-10-PCS codes in Taiwan, set by National Health Insurance Administration. Due to this limitation, some diseases or symptoms in free-text data cannot be recorded in ICD-10 codes. Our model should not only learn the rules about ICD-10 coding rules but also learn NTUH coder’s coding priority. This limitation increases the difficulty when the case contains more than 20 ICD-10-CM codes. The neural network needs to learn the features related to ICD-10 codes and determine whether the code is important enough in the 20 ICD-10 codes limitations.
PCS Results
The complete ICD-10-PCS code has 7 characters and each can be either alpha or numeric. We have 9, 513 labels to predict in our dataset. We use progress, discharge diagnoses and physical examinations as our input data. Unlike ICD-CM’s result, we have the best result F1-score 0.61 when we use 100 as our embedding dimension.
Conclusion
We develop an ICD-10-CM classification model by NLP and deep learning model without any background knowledge from electronic health record data. Previous study [14] focused on ICD-9 classification, where there were 85,522 training samples and the F1-score of 0.41. We observe that ICD classification requires large data for training, and more data can actually help address the data imbalance issue. Furthermore, it may be needed to build a rule-based system for classifying the subtle rules in ICD-10-CM like combination code correctly.
Acknowledgements
This study was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 107-2634-F-002-015). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have declared that no competing interests exist.
REFERENCES
- Chowdhury CG. Natural language processing. Inf Sci Tech. 2003; 37(1): 51-89.
- Pivovarov R, Elhadad N. Automated Methods for the Summarization of Electronic Health Records. J Am Med Inform Assoc. 2015; 22(5):938-947.
- Feller DJ, Zucker J, Yin MT, Gordon P, Elhadad N. Using clinical notes and natural language processing for automated HIV risk assessment. JAIDS. 2018; 77(2): 160-166.
- WHO. 2017; ICD-10 Version (Classification of Diseases).
- Zhang Y, Chen R, Tang J, Sutter WS, Sun J. LEAP: Learning to Prescribe Effective and Safe Treatment Combinations for Multimorbidity. KDD. 2017; 1315-1324.
- Wang Y, Afzal N, Fu S, Wang L, Shen F, Rastegar-Mojarad M, Liu H. MedSTS: A Resource for Clinical Semantic Textual Similarity.2018.
- Farkas R, Szarvas G. Automatic construction of rule-based ICD-9-CM coding systems. BMC bioinformatics. 2018; 9(Suppl 3):S10.
- Loper E, Bird S. NLTK: the natural language toolkit. In Proceedings of the ACL-02 Workshop on Effective tools and methodologies for teaching natural language processing and computational linguistics. ETMTNLP '02. 2002; 1(1): 63-70.
- Mikolov S, Sutskever I, Chen K, Corrado G, Dean J. Distributed Representations of Words and Phrases and their Compositionality. Advanc Neural Inform Proc Sys. 2013; 3111-3119.
- Pennington RJ, Manning C. Glove. Global vectors for word representation: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing EMNLP. 2014; 1532-1543.
- Caruana R, Niculescu-Mizil A. An empirical comparison of supervised learning algorithms. Proceedings of the 23rd International Conference on Machine Learning ICML. 2006; 161-168.
- Chung KCJ, Gulcehre C, Bengio Y. Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling. 2014: 1412.
- Schmidhuber J, Gers FA. LSTM recurrent networks learn simple context free and context sensitive languages. IEEE Transactions on Neural Networks 12. 2001; 6: 1333-1340.
- Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M. Scalable and accurate deep learning for electronic health records. npj Digital Med. 2018; 18: 1-10.